Play all audios:
ABSTRACT The structure of the nuclosome core particle of chromatin in chicken erythrocytes has been examined by using AFM. The 146 bp of DNA wrapped twice around the core histone octamer are
clearly visualized. Both the ends of entry/exit of linker DNA are also demonstrated. The dimension of the nucleosome core particles is ∼ 1-4 nm in height and ∼ 13-22 nm in width. In
addition, superbeads (width of ∼ 48-57 nm, height of ∼ 2-3 nm) are occasionally revealed, two turns of DNA around the core particles are also detected. SIMILAR CONTENT BEING VIEWED BY OTHERS
STRUCTURE OF NATIVE CHROMATIN FIBRES REVEALED BY CRYO-ET IN SITU Article Open access 10 October 2023 LINKER HISTONE DEFINES STRUCTURE AND SELF-ASSOCIATION BEHAVIOUR OF THE 177 BP HUMAN
CHROMATOSOME Article Open access 11 January 2021 ANALYSIS OF THREE-DIMENSIONAL CHROMATIN PACKING DOMAINS BY CHROMATIN SCANNING TRANSMISSION ELECTRON MICROSCOPY (CHROMSTEM) Article Open
access 16 July 2022 INTRODUCTION All eukaryotic chromosomes consists of a regularly repeating protein-DNA complex called the nucleosome (Kornberg, 1977)1. It is traditionally considered to
be the fundamental unit of chromatin structure. The physical properties of nucleosome depend on solution condition such as ionic strength and divalent-ion concentration as well as on
histone-modification state. Recently, the X-ray crystal structure of the nucleosome core particle at 2.8 Å resolution clearly showed how the histone protein octamer was assembled and how 146
bp base pairs of DNA were organized into a superhelix, around it both histone/histone and histone/DNA interactions depended on the histone fold domains (Luger et al 1997)2. Our previous
studies revealed the chromatin folding patterns in chicken erythrocytes by AFM. At the first level of DNA packing, our data showed that an extended beads-on-a-string (width of ∼ 15-20 nm,
height of ∼ 2-3 nm for each individual nucleosome) could be consistently observed. Furthermore, superbeads (width of ∼ 40 nm, height of ∼ 7 nm) arranged with irregular distance along the
extended beads-on-a-string were demonstrated (Qian et al 1997)3. Here, we will show the structure of the nucleosome core particle of chromatin in chicken erythrocytes examined with
tapping-mode scanning force microscopy. The nuclei prepared from chicken erythrocytes were digested with micrococcal nuclease and the samples were then applied to a sepharose 4B column. The
mono-, di- and oligo-nucleosomes were prepared and the linker histone as well as nonhistone proteins were stripped from them (Lutter et al 1978)4. MATERIALS AND METHODS _PREPARATION OF
NUCLEOSOMES FROM CHICKEN ERYTHROCYTES_ Nuclei were prepared from chicken erythrocytes and processed as described by Lutter (1978)4. These nuclei were then digested with 40 units micrococcal
nuclease/ml in the presence of 1 mM-CaCl2 for 5 min at 37°C. The digestion was terminated by addition of Na2EDTA to 2 mM. The nuclei were then centrifuged for 10 min at 5000 g, resuspended
and lysed by pipetting up and down in an equivalent volume of 0.2 mM Na2 EDTA (pH 7.0) and centrifuged again for 10 min at 5000g. The supernatant contains soluble chromatin and 0.127 vol. of
4M-NaCl was added dropwise at 0°C. The sample was then applied to a sepharose 4B column (2 cm × 80 cm) which had been equilibrated with 0.45 M-NaCl, 5 mM-Tris (pH 7.5), 0.2
mM-2-mercaptoethanol. Fractions containing A260 absorbing material were collected. The samples of linker histones-stripped nucleosomes were then diluted to 0.1 M NaCl with 20 mM Tris (pH
7.0) and were used for AFM analysis. _ATOMIC FORCE MICROSCOPY OF NUCLEOSOMES_ For AFM analysis, the mono-, di- and oligo- nucleosomes were deposited on mica which was pretreated with 1% APS
(3-Aminopropyl Triethoxysilane). Excess liquid was blown off with gas. Tapping mode images were obtained on a nanoscope III AFM (Digital instruments, Santa Barbara, CA), using nanoprobe
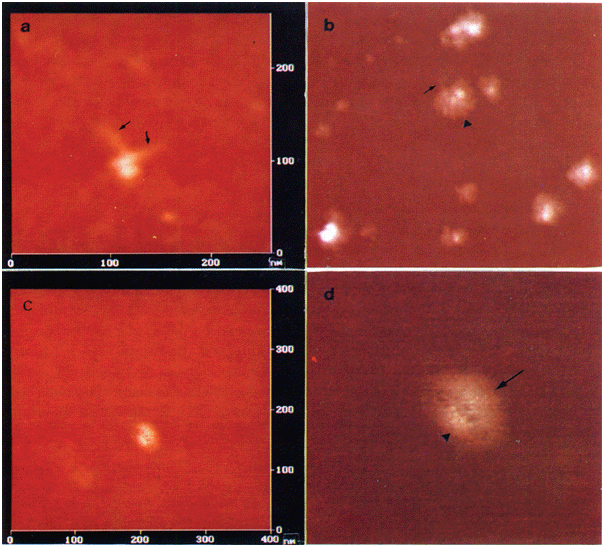
silicon tips, scan rate 1–1.5 HZ. RESULTS _AFM IMAGES OF MONO-NUCLEOSOMES_ In order to gain further insight into the structure of the nucleosome core particle, AFM imaging of native and
linker histone-stripped mono-nucleosomes from chicken erythrocytes was performed. Interestingly, the remarkable features of the nucleosome core particles displayed at different orientation
can be visualized by AFM. The 146 bp of DNA wrapped twice around the core histone octamer are clearly visible and both ends of entry/exit of linker DNA are revealed (Fig 1a). The height of
the nucleosome core particle is ∼ 4 nm and the width of it is ∼ 21 nm. In Fig 1b, another nucleosome core particle wrapped twice by the DNA is also observable (arrow head indicated). Its
both ends of entry/exit of linker DNA can be revealed (arrow indicated). However, The height of this core particle is ∼ 2 nm and its width is ∼ 57 nm. It is difficult to determine whether
this measurement reflects the true dimension of some core particles or if some of the cores are depressed by the force load of the AFM probe. Furthermore, Fig 1c, d. Display a nucleosome
core particle in which two turns of the DNA cann't be visualized owing to the different orientation of the nucleosome core particle. However, both crescent-shaped conformations, which
may consist of core histones, seem to be revealed. Several linkers between the two parts can be also observed (Fig 1d). The height of this nucleosome core particle is ∼ 2 nm and the width of
it is ∼ 20 nm. _VISUALIZATION OF OLIGO-NUCLEOSOMES BY AFM_ The nucleosome core particles in oligo-nucleosomes (lacking linker histones) were also examined by using AFM. Three nucleosomes
are demonstrated in Fig 2a. Two turns of the 146 bp DNA around the core histone octamer can still be visualized, with the dimension of the core particles ∼ 13–17.5 nm in width and ∼ 1–1.6 nm
in height. Fig 2b shows the mono-, di- and oligo-nucleosomes. The 146 bp of DNA wrapped twice around the core histone octamer can be detected in some core particles. The width of core
particles is ∼ 17-20 nm and the height is ∼ 1-2 nm. In addition, some core particles (width of ∼ 43-50 nm, height of ∼ 2-3 nm) can be also revealed (thick arrow indicated in Fig 2b). They
are very similar to the surperbeads as mentioned before (Qian et al 1997)3 Two turns of DNA around these superbeads can be visualized. DISCUSSION For many years, biologists have been trying
to understand how DNA is packaged into chromosomes. At the first level of compaction, the histones and DNA are organized in repeating units called nucleosomes, the histone octamer has been
shown to be internally organized as a tripartite protein assembly (Arents et al 1991)5. A centrally located (H3-H4) tetramer is flanked by two H2A-H2B dimers. It has been hypothesized that
the 146 bp of DNA is wrapped twice around the histone octamer. This complex of the histone octamer and 146 bp of DNA is known as the nucleosome core particle, each nucleosome core particle
is associated with linker histone (H1 or H5). The nucleosome (nucleosome core particle, linker DNA and linker histone) is a fundamental unit of transcriptionally inactive chromatin in all
eukaryotic chromosomes and most of its sequence are in the inactive state for most of time. The nucleosomes (lacking histone H1 and H5) are mobile in physiological condition. Moreover, the
X-ray crystal structure of the complete nucleosome core particle was resolved at 2.8Å resolution (Luger et al 1997)2. The new structure provides a detailed view of the protein and DNA
organization. The overall DNA trajectory approximates 1.65 turns of a superhelix, but the diameter and bending are not uniform. Recently, the atomic force microscopy has been used for
exploring the structure of chromatin (Allen et al 1993 Leuba et al 1998)6, 7. As a useful tool in biological research, AFM can image biological molecules under conditions close to their
native environment. So far it is the only microscope that can achieve nanometer scale resolution on biological samples under native condition. AFM provides more clearly resolved images of
the structure of chromatin than that attained by electron microscopy. In this report, the structure of nucleosome core particle of chromatin in chicken erythrocytes was analyzed by AFM. The
evidence that 146 bp of DNA wrapped twice around the core histone octamer was revealed and both the ends of entry/exit of linker DNA could be clearly visualized. In addition, the
oligo-nucleosomes (lacking linker histones) were also visible with AFM. The height of the nucleosome core particles is ∼ 1-4 nm and the width is 13-22 nm. Furthermore, supper-beads were
occasionally revealed. It seems that the dimension of the nucleosome core particles detected by AFM is larger than that measured by neutron scattering and x-ray diffraction (Imai et al 1986;
Richmond et al 1984)8, 9. Since the present studies were carried out with a tapping mode AFM, in which samples were absorbed to the surface of mica and examined in air at ambient humidity
and very large amount of energy could be deposited into the specimen with each “tap”, the nucleosome core particles might be loosed and their width might. Therefore, be larger than that
measured by other methods. Thus, the improvement of the AFM technique and it's resolution will be undoubtedly essential for the further elucidation of the structure and function of
chromatin. Moreover, the results presented here may provide some new clues to the structure of the native and linker histone-depleted nucleosome core particles prepared from chicken
erythrocytes. REFERENCES * Kornberg RD . Structure of chromatin. _Annu Rev Biochem_ 1977; 46: 931–54. Article CAS Google Scholar * Luger K, AW Mader, RK Richmond, DF Sargent, TJ Richmond
. Crystal structure of the nucleosome core particle at 2.8 ∼ resolution. _Nature_ 1997; 389: 251–60. Article CAS Google Scholar * Qian RL, ZX Liu, MY Zhou, et al. Visualization of
chromatin folding patterns in chicken erythrocytes by atomic force microscopy (AFM). _Cell Research_ 1997; 7: 143–50. Article CAS Google Scholar * Lutter LC . Kinetic analysis of
deoxyribonuclease I cleavages in the nucleosome core: Evidence for a DNA superhelix. _J Mol Biol_ 1978; 124: 391–420. Article CAS Google Scholar * Arents G, RW Burlingame, BC Wang, WE
Love, and EN Moudrianakis, The nucleosomal core histone octamer at 3.1 A resolution: A tripartite protein assembly and a left-handed superhelix. _Proc Natl Acad Sci USA_ 1991; 88: 10148–52.
Article CAS Google Scholar * Allen MJ, XF Dong, TE ONeill et al. Atomic force microscope measurement of nucleosome cores assembled along defined DNA sequences. _Biochemistry_ 1993; 32:
8390–6. Article CAS Google Scholar * Leuba SH, C Bustamante, K Van holde, J Zlatanova . Linker histone tails and N-tails of histone H3 are redundant: Scanning force microscopy studies of
reconstituted fibers. _Biophysical Journal_ 1998; 74: 2830–9. Article CAS Google Scholar * Imai BS, Y Peter, JP Baldwin, RP May, EM Bradbury . Hyperacetylation of core histones does not
cause unfolding of nucleosomes. _J Biol Chem_ 1986; 261: 8784–92. CAS PubMed Google Scholar * Richmond TJ, JT Finch, B Rushton, D Rhodes, A Klug . Structure of the nucleosome core
particle at 7 A resolution. _Nature_ 1984; 311: 532–7. Article CAS Google Scholar Download references ACKNOWLEDGEMENTS This work is supported by the National Natural Science Foundation of
China (Grant No. 39893320) and Foundation of Chinese Academy of Sciences (Grant No. KJ951-A1-603(2)). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Shanghai Institute of Cell Biology,
Chinese Academy of Sciences, Shanghai, 200031, China Hui ZHAO, Shu Bing ZHANG, Chu JIANG, Qi Ye HE & Ruo Lan QIAN * Shanghai Institute of Nuclear Research, Chinese Academy of Sciences,
Shanghai, 201800, China Yi ZHANG & Min Qian LI Authors * Hui ZHAO View author publications You can also search for this author inPubMed Google Scholar * Yi ZHANG View author publications
You can also search for this author inPubMed Google Scholar * Shu Bing ZHANG View author publications You can also search for this author inPubMed Google Scholar * Chu JIANG View author
publications You can also search for this author inPubMed Google Scholar * Qi Ye HE View author publications You can also search for this author inPubMed Google Scholar * Min Qian LI View
author publications You can also search for this author inPubMed Google Scholar * Ruo Lan QIAN View author publications You can also search for this author inPubMed Google Scholar
CORRESPONDING AUTHOR Correspondence to Ruo Lan QIAN. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE ZHAO, H., ZHANG, Y., ZHANG, S. _et al._ The
structure of the nucleosome core particle of chromatin in chicken erythrocytes visualized by using atomic force microscopy. _Cell Res_ 9, 255–260 (1999).
https://doi.org/10.1038/sj.cr.7290024 Download citation * Received: 28 October 1999 * Revised: 12 November 1999 * Accepted: 15 November 1999 * Issue Date: 01 December 1999 * DOI:
https://doi.org/10.1038/sj.cr.7290024 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative KEYWORDS * Nucleosome core particle * chicken erythrocytes *
atomic force microscopy