Play all audios:
ABSTRACT AIM: Blockade of interleukin-6 (IL-6) or its receptor (IL-6R) is effective in preventing the progression of autoimmune diseases, such as systemic lupus erythematosus and rheumatoid
arthritis. In the present study, we established a novel cell-based assay for identifying small molecule IL-6R antagonists. METHODS: HEK293A cells were transfected with recombinant plasmids
pTaglite-SNAP-IL6R and pABhFc-IL6 to obtain membrane-bound IL-6R and recombinant human IL-6 coupled with human Fc fragment (rhIL-6), respectively. A novel screening assay based on the
interaction between IL-6R and rhIL-6 was established, optimized and validated. The stability of the assay was also assessed by calculating the Z′-factor. RESULTS: RhIL-6 dose-dependently
bound to IL-6R expressed at HEK293A cell surface. The IC50 value of the known antagonist ab47215 was 0.38±0.08 μg/mL, which was consistent with that obtained using the traditional method
(0.36±0.14 μg/mL). The value of Z′-factor was 0.68, suggesting that the novel assay was stable for high throughput screening. A total of 474 compounds were screened using the novel screening
assay, and 3 compounds exhibited antagonistic activities (IC50=8.73±0.28, 32.32±9.08, 57.83±4.24 μg/mL). Furthermore, the active compounds dose-dependently inhibited IL-6-induced
proliferation of 7TD1 cells, and reduced IL-6-induced STAT3 phosphorylation in U937 cells. CONCLUSION: A novel cell-based screening assay for identifying small molecule IL-6R antagonists was
established, which simplifies the procedures in traditional cellular ELISA screening and profiling and reduces the costs. SIMILAR CONTENT BEING VIEWED BY OTHERS DISCOVERY OF A SELECTIVE AND
BIOLOGICALLY ACTIVE LOW-MOLECULAR WEIGHT ANTAGONIST OF HUMAN INTERLEUKIN-1Β Article Open access 07 September 2023 CHARACTERISATION OF IL-23 RECEPTOR ANTAGONISTS AND DISEASE RELEVANT MUTANTS
USING FLUORESCENT PROBES Article Open access 19 May 2023 UNVEILING NOVEL INSIGHTS INTO HUMAN IL-6 − IL-6R INTERACTION SITES THROUGH 3D COMPUTER-GUIDED DOCKING AND SYSTEMATIC SITE
MUTAGENESIS Article Open access 07 August 2024 INTRODUCTION Interleukin-6 (IL-6) is a key cytokine that was originally identified as a B-cell differentiation factor in 1985; it is a
multifunctional moderator regulating acute phase reaction, immune response, inflammation, bone metabolism and hematopoiesis1,2. However, under some pathological conditions, continuous
addition of IL-6 has been shown to increase autoantibodies, such as anti-double-stranded DNA (anti-dsDNA) antibody production, making significant contributions to the inflammatory response
and accelerating the progression of autoimmune diseases such as systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA)3,4,5. A blockade of IL-6 or its receptor has been
demonstrated to prevent elevated anti-dsDNA antibody levels and the progression of autoimmune diseases in addition to improving survival rates6. Tocilizumab, a humanized monoclonal antibody
(mAb) against the α-chain of the IL-6 receptor (IL-6Rα, also known as IL-6R), prevents the binding of IL-6 to membrane-bound IL-6R7. The safety and efficacy of tocilizumab has been assessed
in many clinical trials in patients with RA, SLE, juvenile idiopathic arthritis, and Castleman's disease8,9,10. However, therapeutic mAbs are usually produced from non-human cell lines,
and consequently, the exogenous biomacromolecules theoretically can invoke anti-globulin responses in some patients11,12. Therefore, it may be a reasonable strategy to develop small
molecule drugs against IL-6R in the patients with anti-globulin responses to therapeutic mAbs. However, there are currently no small molecule drugs in clinical use or in late clinical trials
that specifically target human IL-6R. For these reasons, research and development of novel IL-6R antagonists with a higher efficacy and less side effects should be of significance in
clinical applications. With a focus on drug discovery, various assays for identifying IL-6R antagonists have been developed during the past several years, which have significantly improved
screening efficiencies. A classical cell-based assay, the 7TD1 cell model, was established by Van Snick in 1986; this assay used an IL-6 dependent mouse-mouse hybrid cell proliferation model
to determine the blocking effects of compounds13. It is an effective method for identifying human IL-6R antagonists and is widely used in targeted drug discovery and fundamental
research14,15. However, its long testing cycle (often 3–4 d) for a proliferation inhibition experiment may be a weakness to rapid screening, and the detection index (cell survival rate)
itself seems to incapably rule out false positives resulting from drug toxicity. With the innovation of the polymerase chain reaction (PCR) technique and optimization of the _Escherichia
coli_ expression system, a screening assay based on phage display technology for identifying IL-6R antagonists was established and applied in the recent past16,17. It is a burgeoning and
efficient screening technique, particularly for identifying candidates with a high affinity. Moreover, the emergence of fully synthetic human phage surface display technology opens up a new
method for reducing immunogenicity18,19. However, there is no doubt that a micro-molecule polypeptide library or a miniaturization antibody library is required; consequently, the novel
screening assay based on phage display technology is merely used for discovering specified types, such as polypeptide drugs, antibodies and other biological agents. Some methods derived from
soluble receptor-ligand binding assays are also widely used in screening IL-6R antagonists20,21. However, each of the assays seemingly has pros and cons. For example, the radioactive
ligand-receptor binding test is considered to be an effective, sensitive, and classical analysis method, but safer methodologies than radiolabeled assays allow the use of radioactivity to be
avoided22. A safe method based on the receptor-ligand binding assay mechanism is the enzyme-linked immunosorbent assay (ELISA); this is a quick, simple and extensive approach21,23. However,
the ELISA still has room for improvement. For example, it has not yet been addressed whether the operation steps can be simplified without influencing detection sensitivity or whether the
binding process of the receptor and its ligand could be similar to the process of the natural membrane-bound receptor binding to its ligand. Therefore, developing novel, effective and
ingenious assays for identifying IL-6R antagonists may provide opportunities to screen for discovering potent, effective antagonists. We report herein an innovative, cell-based screening
assay for identifying IL-6R antagonists by using two new forms of proteins: human membrane IL-6R, located on the HEK293A cell surface, and recombinant human IL-6 (rhIL-6) coupled with the
human Fc fragment. MATERIALS AND METHODS MATERIALS AND INSTRUMENTATION The human IL-6 and IL-6Rα genes were purchased from Origene Technologies. The HEK293A cells, 7TD1 cells and U937 cells
were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The expression plasmid pTaglite-SNAP was purchased from Cisbio Bioassays Corporation. The competent
_Escherichia coli_ cells Top10 were purchased from TransGen Biotech Corporation (Beijing, China). Restriction enzymes, such as _Not_ I and _Xba_ I, were purchased from New England Biolabs.
_Thermus aquaticus_ DNA polymerase and T4 DNA ligase were purchased from Takara Biotech (Dalian, China). RPMI-1640 culture medium, α-MEM culture medium and fetal bovine serum (FBS) were
purchased from Gibco Life Technologies. The transfection reagent Lipofectamine 2000 system was purchased from Invitrogen Corporation. A total of 474 small molecular compounds that belonged
to our in-house library were provided by the National Center for Drug Screening, Institute of Materia Medica, Chinese Academy of Medical Sciences, and each of the compounds was stored in
DMSO and had an initial concentration of 10 mg/mL. The microplate reader, SpectraMax M5, was purchased from BD Biosciences. The nucleus dye 4',6-diamidino-2-phenylindole (DAPI) was
purchased from Partec Flow Cytometry Technology (Görlitz, Germany). The cellomics arrayscan VTI HCS reader was purchased from Thermo Scientific. The cell counting kit-8 (CCK-8) system was
purchased from Dojindo Molecular Technologies Inc (Kyushu, Japan). The Gel DocTM XR+ system and the Quantity One 1-D analysis software were purchased from Bio-Rad Laboratories. All of the
manufacturers' materials and instrumentations described above were obtained from the USA unless otherwise specified. The primary antibodies and secondary antibodies used in this study
and their working concentrations are listed in Table 1, and the DNA primers designed and used in this study are listed in Table 2. CONSTRUCTIONS OF THE RECOMBINANT EUKARYOTIC EXPRESSION
PLASMIDS The full length of the human IL-6Rα chain gene and the IL-6 gene were amplified by PCR using the specific primers listed in Table 2. Using the instrumental enzymes of the gene
engineering mentioned above, the DNA products were reconstructed separately in plasmid pTaglite-SNAP, which is a commercialized vector expressing the receptor of interest at the cell
surface, and in plasmid pABhFc, which is used to express secretory IL-6 coupling with the human Fc fragment at the C-terminus24,25. The recombinants, pTaglite-SNAP-IL6R and pABhFc-IL6, were
analyzed and identified by restriction enzyme digestion and a sequencing analysis. CELL CULTURE AND TREATMENT The HEK293A cells were maintained in α-MEM medium supplemented with 10%
heat-inactivated FBS in a 37 °C incubator with 5% CO2. For all the experiments, the cells were grown to an 80% confluence with no more than 20 passages. The cells were transfected separately
by the expression plasmids pTaglite-SNAP-IL6R and pABhFc-IL6 using the transfection reagent Lipofectamine 2000 system according to the manufacturer's instructions. The 7TD1 cells were
maintained in RPMI-1640 medium supplemented with 10% heat-inactivated FBS and rhIL-6 (4 ng/mL) in a 37 °C incubator with 5% CO2. For the proliferation inhibition test, the cells were washed
three times to remove the growth factor IL-6 and were inoculated in 96-well plates. After being pre-incubated with or without the test compounds for 1 h, the cells were exposed in the
presence or absence of different concentrations of rhIL-6 (or IL-6). The number of viable cells was counted by an automated cell counter every 12 h over 3 d. After 72 h, the viabilities of
the 7TD1 cells were measured by the CCK-8 system26. The U937 cells were maintained in RPMI-1640 medium supplemented with 10% heat-inactivated FBS in a 37 °C incubator with 5% CO2.
IMMUNOFLUORESCENCE ASSAY The expression level of IL-6R was examined by an indirect immunofluorescence assay using a rabbit anti-human IL-6Rα polyclone antibody and its corresponding
secondary antibody. Briefly, the HEK293A cells transfected by the gene pTaglite-SNAP-IL6R for 36 h were washed, fixed with 4% paraformaldehyde for 30 min at room temperature (RT), and
blocked with 2% BSA for 1 h at 37 °C. Next, the cells were incubated with the primary antibody mentioned above for 1 h at 37 °C, followed by the Alexa Fluor® 488-conjugated secondary
antibody. Finally, after being washed, the cells were incubated in DAPI solution for 5 min at RT in the dark. Live cell imaging was simultaneously viewed using a Cellomics arrayscan VTI HCS
reader at an excitation wavelength of 355 nm for DAPI and 495 nm for Alexa Fluor® 488. WESTERN BLOTTING ASSAY The expression level of IL-6R was quantified by a Western blot assay. After
transfection with the gene pTaglite-SNAP-IL6R, the HEK293A cells were collected, and the total protein was extracted in lysis buffer as described by Schreiber _et al_27. A total of 20 μg of
protein extract was resolved on a 10% SDS-PAGE gel, transferred onto a polyvinylidene fluoride membrane, and blocked with 5% BSA in Tris-buffered saline (TBS) for 1 h at 37 °C. The membrane
was washed three times in TBS with 0.1% Tween 20 (TBS-T) and incubated with the anti-IL6Rα polyclone antibody at 4 °C overnight. The membrane was washed three times in TBS-T for 10 min and
incubated with anti-rabbit IgG (HRP conjugate) in TBS. After another three washes with TBS-T for 10 min, the membrane was reacted with an enhanced chemiluminescence system for a moment and
was then exposed to films. The expression level of IL-6R was quantified by scanning densitometry using a Quantity One 1-D analysis system. The levels of the protein signal transducer and the
activator of transcription 3 (STAT3) phosphorylation were also quantified by a Western blot assay. The U937 cells were pre-incubated with or without the test compounds for 1 h and were then
stimulated by rhIL-6 (or IL-6) for 30 min in a 37 °C incubator with 5% CO2. The following procedures for the Western blot assay are similar to those described above except for the primary
antibody involved. PURIFICATION OF RHIL-6 After transfection with the plasmid pABhFc-IL6 for 72 h, the HEK293A cell culture supernatants were collected and filtered with a 0.45-μm filter
membrane. Next, the rhIL-6 in the supernatants was purified by the protein A chromatography method described by Ng _et al_28. The molecular weight and purity of the purified products were
detected by 10% SDS-PAGE. CELLULAR ELISA The direct binding capacity of rhIL-6 coupled with human Fc and the secondary antibody (HRP conjugate) was investigated. Each well of a high-binding
96-microwell plate was coated with different concentrations of rhIL-6 (or bevacizumab or ab47215) for 1 h at 37 °C. The plate was blocked with 2% BSA for 1 h at 37 °C; then, anti-human IgG
goat mAb (HRP conjugate) was added to each well. After three washes with PBS, a colorimetric assay format was followed as described by Emon _et al_29. The absorbance of each well was
determined in a dual-wavelength manner (492–630 nm) using an ELISA reader (Microplate Reader SpectraMax M5). The binding capacity of rhIL-6 coupled with human Fc and the membrane receptor
was investigated. After transfection with plasmid pTaglite-SNAP-IL6R for 36 h, the HEK293A cells in each well were washed and fixed with 4% paraformaldehyde for 30 min at RT and blocked with
2% BSA for 1 h at 37 °C. Next, different concentrations of rhIL-6 were added to the wells, followed by incubation for 1 h at RT. The cells were washed three times to remove the dissociative
rhIL-6. The following procedures for the ELISA were similar to those described in the previous paragraph. Some potential factors, such as temperature states and blocking systems, which may
influence the binding capacity of rhIL-6 and the membrane receptor, were investigated by the cellular ELISA as described above. DETERMINATION OF THE Z′-FACTOR The Z'-factor was
determined to evaluate the stability and suitability of the method in a 96-well format by three separate experiments. To calculate the Z'-factor of the assay, the rhIL-6 reactions
(containing 2% BSA) were assayed in 25 non-marginal wells of a plate; another 25 non-marginal wells of the plate were used for background controls (containing only 2% BSA, assuming that
rhIL-6 was neutralized by the antagonists). Throughout the cell culture assays and cellular ELISA that followed, the same medium or PBS was added to all marginal wells to reduce the errors.
The Z'-factor for the assay was calculated as described by Zhang _et al_30. VERIFICATION OF THE MODIFIED SCREENING ASSAY A known antagonist, ab47215, was used to assess the reliability
of the modified assay31,32. Different concentrations of ab47215 were added to the rhIL-6 reactions and incubated together with membrane IL-6R for 1 h at 37 °C. After three washes with PBS, a
secondary antibody was used to detect the rhIL-6 that was bound to membrane IL-6R as described above. To calculate the block rate (inhibition rate), the mathematical equation was used:
PRELIMINARY SCREENING AND VERIFICATION OF ACTIVE COMPOUNDS A total of 474 compounds from the small molecular compound library were added separately to the rhIL-6 reactions (the final
concentration of each compound was 0.1 mg/mL) for primary screening by the cell-based assay. The positive compounds were re-screened in a secondary screening to test and verify the results.
A previous model described by Vardanyan _et al_21 was used to examine the functions of the active compounds obtained by the preliminary screening and thus to assess the reliability of the
novel assay (several negative compounds in the preliminary screening were included as a control). The procedure is as follows: the plates were coated with soluble IL-6R overnight at 4 °C and
pre-blocked with 1% BSA at RT for 1 h. After three washes, the wells were incubated with human IL-6 plus one of eight concentrations of the compounds at RT for 2 h. After washing to remove
any unbound IL-6, the wells were incubated with an anti-human IL-6 antibody (primary antibody) at 37 °C for 1 h, and washed and incubated with a secondary antibody (HRP conjugate) at 37 °C
for 30 min. The IC50 of each active compound was calculated using Eq (1) described above. STATISTICAL ANALYSIS The results are expressed as the mean±SEM of at least three independent
experiments. The statistical significance was assessed by Student's _t_-test for paired populations or a one-way ANOVA followed by an appropriate _post hoc_ test. _P_ values <0.05
were considered statistically significant. RESULTS CONSTRUCTION OF THE RECOMBINANT EUKARYOTIC EXPRESSION PLASMIDS We initially amplified the IL-6Rα and IL-6 genes by PCR using two pairs of
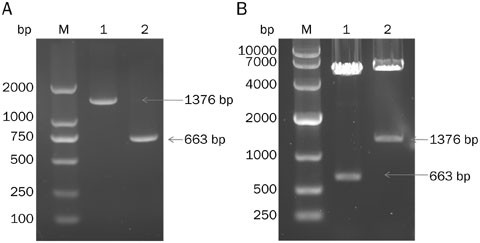
specific primers listed in Table 1. One percent agarose gel electrophoresis was used to detect the relative molecular weights of the amplification products. As shown in Figure 1A, the
electrophoresis band analysis revealed that the DNA products were 1000–2000 base pairs (bp) in length and 500–750 bp in length, respectively, which was in agreement with the theoretical
molecular weights of the IL-6Rα and IL-6 genes33,34. Similarly, we detected the validities of the recombinant plasmids pTaglite-SNAP-IL6R and pABhFc-IL6 with an electrophoresis analysis
after the accomplishment of gene recombination (Figure 1B). The results of this electrophoresis analysis and the DNA sequencing analysis (data not shown) confirmed that the human
IL-6Rα-chain and IL-6 genes had been cloned correctly into the eukaryotic expression vectors. EXPRESSION OF MEMBRANE IL-6R After transfection with pTaglite-SNAP-IL6R for 36 h, the expression
level of IL-6R was investigated by an immunofluorescence assay. As shown in Figure 2A, the transfected cell surface could emit a forceful green fluorescence, which suggested that plentiful
receptors existed at the cell surface compared with the normal control group and the control plasmid group. The expression level of IL-6R was quantified by a Western blot assay which had a
similar result as the immunofluorescence assay. Compared to normal cells and cells that were transfected by the control plasmid pTaglite-SNAP (blank load), the level of IL-6R significantly
increased (Figure 2B). The results above demonstrated that IL-6R was successfully expressed and oriented at the HEK293A cell surface. SECRETION EXPRESSION AND ACTIVITY IDENTIFICATION OF
RHIL-6 The plasmid pABhFc was an engineering vector that was used for expressing secretory human IgG heavy chains that were constructed by our laboratory19,25. Based on the characteristics
of the plasmid, we obtained soluble recombinant IL-6, which is a homodimer protein (purity >90% by SDS-PAGE), as shown in Figure 3A. After the disulfide bond was broken by reduction
loading buffer, rhIL-6 assumed a monomer form with a 50 kDa relative molecular weight, which was consistent with the expected results. To assess the functions of rhIL-6, we investigated its
biological activity using the 7TD1 cell line, which is an IL6-dependent mouse cell. The number of 7TD1 cells ceased to increase at 36 h in the absence of rhIL-6. However, the number of 7TD1
cells in the presence of rhIL-6 (even at a low concentration of 1 ng/mL) continuously increased until the nutrients were depleted at 72 h (Figure 3B). The results of the CCK-8 assay showed
that cell viability declined in a dose-dependent manner with decreasing concentrations of rhIL-6 (Figure 3C), which was in agreement with that of the above cell number count analysis. After
being stimulated by different concentrations of rhIL-6 for 30 min, the level of protein STAT3 phosphorylation in human U937 cells significantly increased compared to the control group
(Figure 3D). Phosphorylated STAT3 (p-STAT3) is a crucial signal molecule in the JAK/STAT signaling pathway that is mediated by IL-635,36. The binding of IL-6 to membrane IL-6R induces
phosphorylation of STAT3 at Y705 residues, and p-STAT3 dimerizes and translocates to the nucleus, where it binds to the promoter of target genes to regulate transcription35,37. Our results
here indicated that rhIL-6 was able to bind to natural human IL-6R and then mediate signal transduction. All the findings using the 7TD1 and U937 cells above confirmed that the homodimer
protein rhIL-6 coupled with the human Fc fragment showed a bioactivity similar to that of natural IL-6. ESTABLISHMENT OF A SCREENING ASSAY BASED ON A CELLULAR ELISA We initially determined
the usefulness of the Fc fragment of rhIL-6 by using a simple and easy ELISA. As shown in Figure 4A, the absorbance value increased with the rise of the concentrations of rhIL-6, indicating
that rhIL-6 could bind to anti-human IgG goat mAb in a dose-dependent manner, consistent with the positive control bevacizumab (a human IgG-targeted VEGF). However, the negative control
ab47215, which was a mouse mAb that did not contain the human Fc fragment, could not bind to the secondary antibody. These results provide a foundation for the establishment of a simple and
convenient cellular ELISA. Next, a preliminary screening assay based on the cellular ELISA was tentatively established after the HEK293A cells were transfected by plasmid pTaglite-SNAP-IL6R.
The absorbance value increased with the increasing concentrations of rhIL-6 (Figure 4B), demonstrating that rhIL-6 could bind to IL-6R expressed at the HEK293A cell surface in a
dose-dependent manner. Finally, some potential factors influencing the capacity of rhIL-6 binding to the membrane receptor were investigated. We found that the capacity showed significant
differences in different temperature states (Figure 4C), and the number of rhIL-6 binding to membrane receptors reached saturation in the first 10 min at 37 °C, indicating that 37 °C might
be the optimum temperature for a rapid screening. Some common blocking systems for immunological tests, such as 2% BSA, 0.2% casein and 5% milk, seemingly had no obvious effect on the
binding capacity (Figure 4D). ASSESSMENTS OF STABILITY AND RELIABILITY After optimizing these procedures, we determined the Z'-factor of the assay and used a concentration of 10 μg/mL
of rhIL-6 as a final concentration according to the results of Figure 4B. Under this circumstance, the wells coated with transfected cells presented intense absorbance values compared with
the control wells, which were also coated with transfected cells but did not contain rhIL-6 (Figure 5A). Using a 96-well format, we were able to screen for IL-6R antagonists with a
Z'-factor of 0.68. The mouse antibody ab47215, which can competitively antagonize IL-6R, was used to evaluate the reliability of the assay. As shown in Figure 5B, ab47215 could block
rhIL-6 from binding to its membrane receptor in a dose-dependent manner in this novel screening assay. The IC50 of ab47215 was 0.38±0.08 μg/mL, which was consistent with that obtained from
the traditional model (IC50 =0.36±0.14 μg/mL). In addition, Figure 5B also shows a comparison of the assay with traditional assays in terms of maximum inhibitory rates converted from signal
values. The maximum inhibitory rate in the new method was slightly lower than that in the traditional method at the same concentration of ab47215. These results demonstrated that our assay
reported herein could identify known IL-6R antagonists. APPLICATION AND EVALUATION OF THE NOVEL ASSAY A total of 474 compounds were screened at random, as shown in Figure 6A, and ten of
those compounds showed some antagonistic activities against membrane IL-6R. After a secondary screening (re-screening, data not shown), the antagonistic activities of three active compounds
were confirmed. The former two of the three have the same basic parent structure as 7-(methylsulfonyl)-4-oxo-3,4-dihydropyrazolo[5,1-_d_][1,2,3,5]tetrazine, and the latter compound is a
derivative of tetrahydrofuro[3,2-c]naphtho[2,1-e]oxepine-1,3-dione. The other model, which was used to evaluate the functions of the three active compounds and the reliability of the novel
assay, illuminated that the novel screening assay described in this study was reliable. As shown in Figure 6B, the active compounds were able to block soluble IL-6R from binding to its
ligand in a dose-dependent manner. The IC50 of the compounds were 32.32±9.08, 8.73±0.28 and 57.83±4.24 μg/mL, respectively. The 95% confidence intervals for the mean values were
approximately 23.74 to 44.0, 5.69 to 13.38 and 42.79 to 78.16 μg/mL, respectively. However, several negative compounds in the preliminary screening were unable to show obvious antagonistic
activities (data not shown). In addition, further evaluation showed that the active compounds were able to inhibit the proliferation of the 7TD1 cells induced by IL-6 in a dose-dependent
manner (Figure 6C) and reduced the phosphorylation level of STAT3 in the U937 cells induced by IL-6 (Figure 6D). These results demonstrated that the active compounds were antagonists against
IL-6R, and the assay that we established was reliable for identifying IL-6R antagonists. DISCUSSION In summary, an innovative cell-based screening assay was established by preparing two new
forms of proteins, human membrane-bound IL-6R and rhIL-6 coupled with the human Fc fragment, which were obtained separately by transfecting two novel recombinant eukaryotic expression
plasmids, pTaglite-SNAP-IL6R and pABhFc-IL6, into HEK293A cells. The assay was stable and reliable, which was evaluated by applying the Z'-factor and confirmed by utilizing a known
antagonist and an existing model. There were two advantages to this assay. First, the orientation expression of IL-6R at the HEK293A cell surface and the analogous expression patterns
targeting IL-6R have not been reported. The natural membrane-bound IL-6R is now considered to play a key role in the physiological and pathological progress, and consequently, compared with
the classical soluble receptor-ligand binding assay at the biochemical level, this novel assay that is conducted at the cell level might make the process of IL-6 binding to IL-6R expressed
at the cell surface more similar to the natural binding process. Second, rhIL-6, which is a new form of recombinant protein coupled with the human Fc fragment, could directly bind to a
secondary antibody, anti-human IgG goat mAb. However, the classical ELISA usually requires an anti-IL6 or IL-6R antibody as a primary antibody followed by a corresponding secondary antibody
(for instance, HRP conjugate). The innovative fusion expression of rhIL-6 in the study played the function of the primary antibody and thus prevented the tedious process necessary in the
ELISA without any obvious interference. It is well-known that commercial primary antibodies are expensive, especially under the conditions of large-scale use in high throughput screening.
Therefore, the new assay in this study simplified the procedures and reduced costs compared with the classical cellular ELISA19. Furthermore, based on the functional performance, rhIL-6
would be appropriate for the soluble IL-6R binding assay and would be able to simplify the steps. In conclusion, we established and applied an innovative cell-based screening assay for
identifying IL-6R antagonists, which is a new and relatively easy method that provides a fresh strategy for other similar receptor-ligand binding assays. AUTHOR CONTRIBUTION Yang-yang HE
designed the experiments and drafted the manuscript; Yu YAN and Chang ZHANG carried out the cytological experiments, signaling studies and verification experiment of the assay; Peng-yuan LI
and Ping WU performed the studies of eukaryotic expression of the membrane receptor and its ligand; Peng DU participated in the purification of rhIL-6; Da-di ZENG and Jian-song FANG carried
out the statistical analysis; Guan-hua DU and Shuang WANG performed the cellular ELISA, the evaluation of the assay, participated in the experimental design and drafted the manuscript.
REFERENCES * Hirano T . Interleukin 6 (IL-6) and its receptor: their role in plasma cell neoplasias. _Int J Cell Cloning_ 1991; 9: 166–84. Article CAS Google Scholar * Mi XB, Zeng FQ .
Hypomethylation of interleukin-4 and -6 promoters in T cells from systemic lupus erythematosus patients. _Acta Pharmacol Sin_ 2008; 29: 105–12. Article CAS Google Scholar * Pelliniemi TT,
Irjala K, Mattila K, Pulkki K, Rajamaki A, Tienhaara A, _et al_. Immunoreactive interleukin-6 and acute phase proteins as prognostic factors in multiple myeloma. Finnish Leukemia Group.
_Blood_ 1995; 85: 765–71. CAS PubMed Google Scholar * Kishimoto T . The biology of interleukin-6. _Blood_ 1989; 74: 1–10. CAS PubMed Google Scholar * Wei ZF, Jiao XL, Wang T, Lu Q, Xia
YF, Wang ZT, _et al_. Norisoboldine alleviates joint destruction in rats with adjuvant-induced arthritis by reducing RANKL, IL-6, PGE2, and MMP-13 expression. _Acta Pharmacol Sin_ 2013; 34:
403–13. Article CAS Google Scholar * Liang B, Gardner DB, Griswold DE, Bugelski PJ, Song XYR . Anti-interleukin-6 monoclonal antibody inhibits autoimmune responses in a murine model of
systemic lupus erythematosus. _Immunology_ 2006; 119: 296–305. Article CAS Google Scholar * Nishimoto N, Kishimoto T . Humanized antihuman IL-6 receptor antibody, tocilizumab. _Handb Exp
Pharmacol_ 2008; 181: 151–60. Article CAS Google Scholar * Straub RH, Härle P, Yamana S, Matsuda T, Takasugi K, Kishimoto T, _et al_. Anti-interleukin-6 receptor antibody therapy favors
adrenal androgen secretion in patients with rheumatoid arthritis: A randomized, double-blind, placebo-controlled study. _Arthritis Rheum_ 2006; 54: 1778–85. Article CAS Google Scholar *
Maeshima K, Ishii K, Torigoe M, Imada C, Iwakura M, Hamasaki H, _et al_. Successful tocilizumab and tacrolimus treatment in a patient with rheumatoid arthritis complicated by systemic lupus
erythematosus. _Lupus_ 2012; 21: 1003–6. Article CAS Google Scholar * Bykerk VP, Ostör AJ, Alvaro-Gracia J, Pavelka K, Ivorra JA, Graninger W, _et al_. Tocilizumab in patients with active
rheumatoid arthritis and inadequate responses to DMARDs and/or TNF inhibitors: a large, open-label study close to clinical practice. _Ann Rheum Dis_ 2012; 71: 1950–4. Article CAS Google
Scholar * Isaacs JD . Antibody engineering to develop new antirheumatic therapies. _Arthritis Res Ther_ 2009; 11: 225. Article Google Scholar * Zhao L, Ren TH, Wang DD . Clinical
pharmacology considerations in biologics development. _Acta Pharmacol Sin_ 2012; 33: 1339–47. Article CAS Google Scholar * Van Snick J, Cayphas S, Vink A, Uyttenhove C, Coulie PG, Rubira
MR, _et al_. Purification and NH2-terminal amino acid sequence of a T-cell-derived lymphokine with growth factor activity for B-cell hybridomas. _Proc Natl Acad Sci U S A_ 1986; 83: 9679–83.
Article CAS Google Scholar * Ward LD, Hammacher A, Zhang JG, Morton CJ, Simpson RJ, Weinstock J, _et al_. Role of the C-terminus in the activity, conformation, and stability of
interleukin-6. _Protein Sci_ 1993; 2: 1472–81. Article CAS Google Scholar * Hayashi M, Rho MC, Fukami A, Enomoto A, Nonaka S, Sekiguchi Y, _et al_. Biological activity of a novel
nonpeptide antagonist to the interleukin-6 receptor 20S, 21-epoxy-resibufogenin-3-formate. _J Pharmacol Exp Ther_ 2002; 303: 104–9. Article CAS Google Scholar * Naimuddin M, Kobayashi S,
Tsutsui C, Machida M, Nemoto N, Sakai T, _et al_. Directed evolution of a three-finger neurotoxin by using cDNA display yields antagonists as well as agonists of interleukin-6 receptor
signaling. _Mol Brain_ 2011; 4: 1–16. Article Google Scholar * Yamaguchi J, Naimuddin M, Biyani M, Sasaki T, Machida M, Kubo T, _et al_. cDNA display: a novel screening method for
functional disulfide-rich peptides by solid-phase synthesis and stabilization of mRNA-protein fusions. _Nucleic Acids Res_ 2009; 37: e108. Article Google Scholar * Huang JX, Bishop-Hurley
SL, Cooper MA . Development of anti-infectives using phage display: biological agents against bacteria, viruses, and parasites. _Antimicrob Agents Chemother_ 2012; 56: 4569–82. Article CAS
Google Scholar * Yu R, Wang S, Yu YZ, Du WS, Yang F, Yu WY, _et al_. Neutralizing antibodies of botulinum neurotoxin serotype A screened from a fully synthetic human antibody phage
display library. _J Biomol Screen_ 2009; 14: 991–8. Article CAS Google Scholar * Naruishi K, Takashiba S, Chou HH, Arai H, Nishimura F, Murayama Y . Role of soluble interleukin-6 receptor
in inflamed gingiva for binding of interleukin-6 to gingival fibroblasts. _J Periodontal Res_ 1999; 34: 296–300. Article CAS Google Scholar * Vardanyan M, Melemedjian OK, Price TJ,
Ossipov MH, Lai J, Roberts E, _et al_. Reversal of pancreatitis-induced pain by an orally available, small molecule interleukin-6 receptor antagonist. _Pain_ 2010; 151: 257–65. Article CAS
Google Scholar * Weiergräber O, Schneider-Mergener J, Grötzinger J, Wollmer A, Küster A, Exner M, _et al_. Use of immobilized synthetic peptides for the identification of contact sites
between human interleukin-6 and its receptor. _FEBS Lett_ 1996; 379: 122–6. Article Google Scholar * Ueda O, Tateishi H, Higuchi Y, Fujii E, Kato A, Kawase Y, _et al_. Novel
genetically-humanized mouse model established to evaluate efficacy of therapeutic agents to human interleukin-6 receptor. _Sci Rep_ 2013; 3: 1196. Article Google Scholar * Zwier JM, Roux
T, Cottet M, Durroux T, Douzon S, Bdioui S, _et al_. A fluorescent ligand-binding alternative using Tag-lite® technology. _J Biomol Screen_ 2010; 15: 1248–59. Article CAS Google Scholar *
Zhang C, Tang P, He Y, Du P, Sun ZW, Wang S, _et al_. Secretion expression of extracellular region of EGFR using mammalian cell and its identification. _Lett Biotechnol_ 2012; 23: 635–9.
CAS Google Scholar * Dai Y, Chen J, Li H, Li S, Chen J, Ding Y, _et al_. Characterizing the effects of VPA, VC and RCCS on rabbit keratocytes onto decellularized bovine cornea. _PLoS One_
2012; 7: e 50114. Article CAS Google Scholar * Schreiber E, Harshman K, Kemler I, Malipiero U, Schaffner W, Fontana A . Astrocytes and glioblastoma cells express novel octamer-DNA binding
proteins distinct from the ubiquitous Oct-1 and B cell type Oct-2 proteins. _Nucleic Acids Res_ 1990; 18: 5495–503. Article CAS Google Scholar * Ng CK, Osuna-Sanchez H, Valéry E,
Sørensen E, Bracewell DG . Design of high productivity antibody capture by protein A chromatography using an integrated experimental and modeling approach. _J Chromatogr B_ 2012; 899:
116–26. Article CAS Google Scholar * Emon JMV, Chuang JC, Lordo RA, Schrock ME, Nichkova M, Gee SJ, _et al_. An enzyme-linked immunosorbent assay for the determination of dioxins in
contaminated sediment and soil samples. _Chemosphere_ 2008; 72: 95–103. Article Google Scholar * Zhang J, Chung TD, Oldenburg KR . Validation of high throughput screening assays. _J Biomol
Screen_ 1999; 4: 67–73. Article CAS Google Scholar * Touboul C, Lis R, Al Farsi H, Raynaud CM, Warfa M, Althawadi H, _et al_. Mesenchymal stem cells enhance ovarian cancer cell
infiltration through IL6 secretion in an amniochorionic membrane based 3D model. _J Transl Med_ 2013; 11: 28. Article CAS Google Scholar * Che Q, Liu BY, Liao Y, Zhang HJ, Yang TT, He YY,
_et al_. Activation of a positive feedback loop involving IL-6 and aromatase promotes intratumoral 17β-estradiol biosynthesis in endometrial carcinoma microenvironment. _Int J Cancer_ 2014;
135: 282–94. Article CAS Google Scholar * Yamasaki K, Taga T, Hirata Y, Yawata H, Kawanishi Y, Seed B, _et al_. Cloning and expression of the human interleukin-6 (BSF-2/IFN beta 2)
receptor. _Science_ 1988; 241: 825–8. Article CAS Google Scholar * May LT, Shaw JE, Khanna AK, Zabriskie JB, Sehgal PB . Marked cell-type-specific differences in glycosylation of human
interleukin-6. _Cytokine_ 1991; 3: 204–11. Article CAS Google Scholar * Heinrich P, Behrmann I, Muller-Newen G, Schaper F, Graeve L . Interleukin-6-type cytokine signalling through the
gp130/Jak/STAT pathway1. _Biochem J_ 1998; 334: 297–314. Article CAS Google Scholar * Liu DB, Hu GY, Long GX, Qiu H, Mei Q, Hu GQ . Celecoxib induces apoptosis and cell-cycle arrest in
nasopharyngeal carcinoma cell lines via inhibition of STAT3 phosphorylation. _Acta Pharmacol Sin_ 2012; 33: 682–90. Article CAS Google Scholar * Jung JE, Kim GS, Chan PH . Neuroprotection
by interleukin-6 is mediated by signal transducer and activator of transcription 3 and antioxidative signaling in ischemic stroke. _Stroke_ 2011; 42: 3574–9. Article Google Scholar
Download references ACKNOWLEDGEMENTS This study was supported by the National Scientific & Technological Major Project for “Significant New Drugs Creation” (No 2012ZX09103-101-078) and
the Special Foundation for Scientific Research in Public Health Industry (No 200902008) and the Doctoral Innovation Foundation (No 521005-31034). We are grateful to Dr Xiu-ying YANG (Chinese
Academy of Medical Sciences & Peking Union Medical College) for the introduction and the suggestion of constructing the plasmid pTaglite-SNAP-IL6R. AUTHOR INFORMATION AUTHORS AND
AFFILIATIONS * Beijing Key Laboratory of Drug Targets Identification and Drug Screening, Institute of Materia Medica, Chinese Academy of Medical Sciences and Peking Union Medical College,
Beijing, 100050, China Yang-yang He, Yu Yan, Ping Wu, Jian-song Fang & Guan-hua Du * Institute of Biotechnology, Chinese Academy of Military Medical Sciences, Beijing, 100071, China
Chang Zhang, Peng Du, Da-di Zeng & Shuang Wang * Drug Manufacturing Room of Pharmaceutical Department, Henan Provincial People's Hospital, Zhengzhou, 450003, China Peng-yuan Li
Authors * Yang-yang He View author publications You can also search for this author inPubMed Google Scholar * Yu Yan View author publications You can also search for this author inPubMed
Google Scholar * Chang Zhang View author publications You can also search for this author inPubMed Google Scholar * Peng-yuan Li View author publications You can also search for this author
inPubMed Google Scholar * Ping Wu View author publications You can also search for this author inPubMed Google Scholar * Peng Du View author publications You can also search for this author
inPubMed Google Scholar * Da-di Zeng View author publications You can also search for this author inPubMed Google Scholar * Jian-song Fang View author publications You can also search for
this author inPubMed Google Scholar * Shuang Wang View author publications You can also search for this author inPubMed Google Scholar * Guan-hua Du View author publications You can also
search for this author inPubMed Google Scholar CORRESPONDING AUTHORS Correspondence to Shuang Wang or Guan-hua Du. POWERPOINT SLIDES POWERPOINT SLIDE FOR FIG. 1 POWERPOINT SLIDE FOR FIG. 2
POWERPOINT SLIDE FOR FIG. 3 POWERPOINT SLIDE FOR FIG. 4 POWERPOINT SLIDE FOR FIG. 5 POWERPOINT SLIDE FOR FIG. 6 RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE He, Yy., Yan, Y., Zhang, C. _et al._ Establishment of a novel cell-based assay for screening small molecule antagonists of human interleukin-6 receptor. _Acta Pharmacol Sin_ 35,
1453–1462 (2014). https://doi.org/10.1038/aps.2014.90 Download citation * Received: 07 May 2014 * Accepted: 18 July 2014 * Published: 27 October 2014 * Issue Date: November 2014 * DOI:
https://doi.org/10.1038/aps.2014.90 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently
available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative KEYWORDS * IL-6 * IL-6 receptor * screening assay * high throughput
screening * eukaryotic expression * cellular ELISA * autoimmune disease