Play all audios:
The presence of functional regulatory polymorphism at the interleukin 6 (IL6) locus is uncertain, with many conflicting in vitro findings. To examine the in vivo effect of the three putative
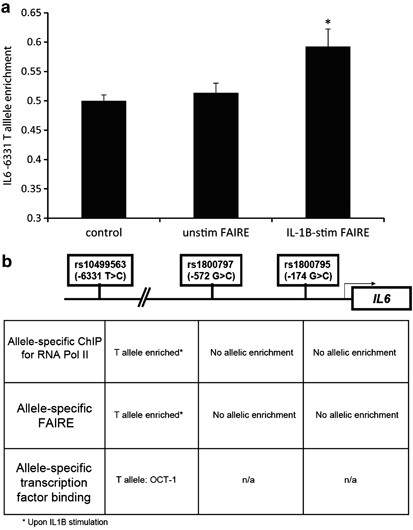
functional IL6 promoter variants, −174G>C, −572G>C and −6331T>C, two complementary techniques, allele-specific chromatin immunoprecipitation and allele-specific formaldehyde-assisted
isolation of regulatory elements, were carried out using unrelated lymphoblast cell lines of known genotype. There were no allele-specific effects for all three single-nucleotide
polymorphisms (SNPs) under basal conditions. Upon IL-1β stimulation, however, allele-specific effects were seen for the −6331 allele, which showed both increased RNA polymerase II loading
(56%, P=0.001) and increased open chromatin (59%, P=0.004) for the T allele, which is in line with previous reports of this SNP and the effects from acute inflammation. These studies
highlight the importance of examining chromatin under different environmental conditions when studying the functionality of regulatory polymorphisms.
This work was supported by the British Heart Foundation (RG05/014 and PG08/008), and Arthritis Research UK (programme grant no. 17287). We thank Winship Herr for providing the Oct-1
expression plasmid.
Centre for Cardiovascular Genetics, UCL Institute of Cardiovascular Science, University College London, London, UK
Division of Infection & Immunity, Centre for Paediatric and Adolescent Rheumatology, University College London, London, UK
Supplementary Information accompanies the paper on Genes and Immunity website
Anyone you share the following link with will be able to read this content: