Play all audios:
Whole-genome duplication followed by massive gene loss and specialization has long been postulated as a powerful mechanism of evolutionary innovation. Recently, it has become possible to
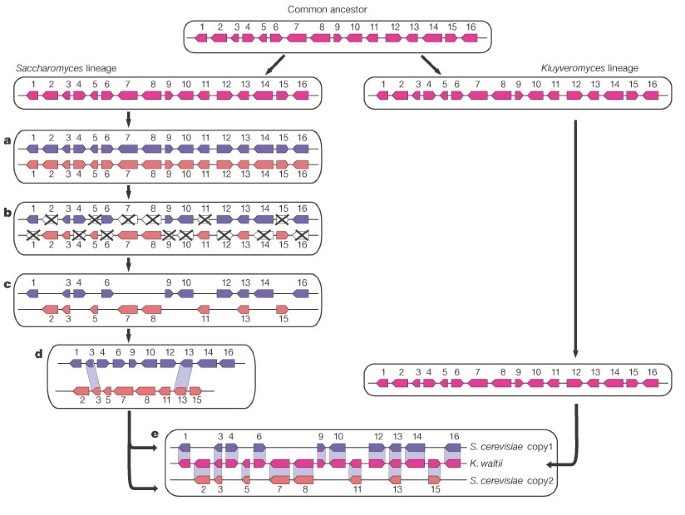
test this notion by searching complete genome sequence for signs of ancient duplication. Here, we show that the yeast Saccharomyces cerevisiae arose from ancient whole-genome duplication, by
sequencing and analysing Kluyveromyces waltii, a related yeast species that diverged before the duplication. The two genomes are related by a 1:2 mapping, with each region of K. waltii
corresponding to two regions of S. cerevisiae, as expected for whole-genome duplication. This resolves the long-standing controversy on the ancestry of the yeast genome, and makes it
possible to study the fate of duplicated genes directly. Strikingly, 95% of cases of accelerated evolution involve only one member of a gene pair, providing strong support for a specific
model of evolution, and allowing us to distinguish ancestral and derived functions.
We thank I. Roberts for providing strains; M. Endrizzi, L.-J. Ma, D. Qi and the staff of the MIT/Whitehead Institute Center for Genome Research who generated the shotgun sequence from
Kluyveromyces waltii; J. Butler and the Arachne assembly team who generated the genome assembly; D. Bartel, S. Calvo, J. Galagan, D. Jaffe and J. Vinson for discussions and comments on the
manuscript; and G. Fink and A. Murray for discussions and advice.
The Broad Institute, Massachusetts Institute of Technology and Harvard University, Cambridge, Massachusetts, 02138, USA
MIT Computer Science and Artificial Intelligence Laboratory, Cambridge, Massachusetts, 02139, USA
Whitehead Institute for Biomedical Research, Cambridge, Massachusetts, 02139, USA
The authors declare that they have no competing financial interests.
Assembly_stats.xls; 16 kb; Description of assembly, scaffolds, contigs, and chromosomes
Kwal_contigs.fasta; 10,797 kb; The raw nucleotide sequence of all contigs
Kwal_scaffolds.txt; 14 kb; Scaffolds (supercontigs) giving relative order, orientation, and spacing of contigs
predicted_ORFs.fasta; 7,459 kb; Raw nucleotide sequence of all predicted Open Reading Frames (ORFs)
predicted_proteins.fasta; 2,528 kb; Protein translation of all Open Reading Frames (ORFs)
Kwal_forward.txt; 993 kb; Annotation of each K. waltii gene and its best match to S. cerevisiae (forward: Kwal => Scer)
Kwal_forward_full.txt; 1,482 kb; Annotation of each K. waltii gene and all its matches to S. cerevisiae (forward: Kwal => Scer)
Kwal_homology_groups_large.txt; 105 kb; Gene families (groups of genes with ambiguous correspondence) that contain at least 4 members
Kwal_homology_groups_small.txt; 182 kb; Gene families (groups of genes with ambiguous correspondence) that contain 3 members of fewer
Kwal_reverse.txt; 776 kb; Coordinates of each S. cerevisiae gene and its best match in K. waltii (reverse: Kwal ≤ Scer)
Kwal_reverse_full.txt; 1,086 kb; Coordinates of each S. cerevisiae gene and all its matches in K. waltii (reverse: Kwal ≤ Scer)
Kwal_synteny_blocks.txt; 28 kb; Blocks of conserved gene order (synteny) between the two species
Matches_by_chromosome.xls; 420 kb; Gene correspondence, grouped by chromosome (shows duplicate mapping)
Matches_by_chromosome_with_syn.xls; 480 kb; Gene correspondence, grouped by chromosome with synteny block information
Paired_regions.xls; 100 kb; Paired regions in S. cerevisiae as defined by blocks of doubly conserved synteny (DCS)
all_scaffolds.pdf; 1,200 kb; Detailed visualization of gene correspondence for every K. waltii chromosome
poster.pdf; 1,127 kb; Detailed visualization of gene correspondence for all K. waltii chromosomes (single page)
all_tiles.html; 257 kb; Detailed visualization of paired sister regions interleaved with K. waltii in 253 DCS blocks
tile_000.jpg; 71 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_001.jpg; 74 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_002.jpg; 120 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_003.jpg; 119 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_004.jpg; 122 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_005.jpg; 113 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_006.jpg; 120 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_007.jpg; 119 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_008.jpg; 104 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_009.jpg; 107 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_010.jpg; 129 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_011.jpg; 78 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_012.jpg; 52 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_013.jpg; 125 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_014.jpg; 105 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_015.jpg; 115 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_016.jpg; 122 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_017.jpg; 73 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_018.jpg; 77 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_019.jpg; 129 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_020.jpg; 38 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_021.jpg; 71 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_022.jpg; 128 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_023.jpg; 107 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_024.jpg; 97 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_025.jpg; 57 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_250.jpg; 79 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_251.jpg; 115 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
tile_252.jpg; 80 kb; Tiling of K. waltii genome in duplicate regions. See all_tiles.html for index of genes in duplicate regions
scer_pairs_01.pdf; 9 kb; S. cerevisiae chromosome 1, connected with its sister regions in S. cerevisiae chromosomes
scer_pairs_02.pdf; 12 kb; S. cerevisiae chromosome 2, connected with its sister regions in S. cerevisiae chromosomes
scer_pairs_03.pdf; 11 kb; S. cerevisiae chromosome 3, connected with its sister regions in S. cerevisiae chromosomes
scer_pairs_04.pdf; 14 kb; S. cerevisiae chromosome 4, connected with its sister regions in S. cerevisiae chromosomes
scer_pairs_05.pdf; 12 kb; S. cerevisiae chromosome 5, connected with its sister regions in S. cerevisiae chromosomes
scer_pairs_06.pdf; 11 kb; S. cerevisiae chromosome 6, connected with its sister regions in S. cerevisiae chromosomes
scer_pairs_07.pdf; 14 kb; S. cerevisiae chromosome 7, connected with its sister regions in S. cerevisiae chromosomes
scer_pairs_08.pdf; 12 kb; S. cerevisiae chromosome 8, connected with its sister regions in S. cerevisiae chromosomes
scer_pairs_09.pdf; 12 kb; S. cerevisiae chromosome 9, connected with its sister regions in S. cerevisiae chromosomes
scer_pairs_10.pdf; 13 kb; S. cerevisiae chromosome 10, connected with its sister regions in S. cerevisiae chromosomes
scer_pairs_11.pdf; 12 kb; S. cerevisiae chromosome 11, connected with its sister regions in S. cerevisiae chromosomes
scer_pairs_12.pdf; 14 kb; S. cerevisiae chromosome 12, connected with its sister regions in S. cerevisiae chromosomes
scer_pairs_13.pdf; 13 kb; S. cerevisiae chromosome 13, connected with its sister regions in S. cerevisiae chromosomes
scer_pairs_14.pdf; 12 kb; S. cerevisiae chromosome 14, connected with its sister regions in S. cerevisiae chromosomes
scer_pairs_15.pdf; 13 kb; S. cerevisiae chromosome 15, connected with its sister regions in S. cerevisiae chromosomes
scer_pairs_16.pdf; 13 kb; S. cerevisiae chromosome 16, connected with its sister regions in S. cerevisiae chromosomes
Browse.html; 141 kb; Browse three-way nucleotide/protein/translation alignments for all 457 gene pairs
translate.tgz; 961 kb; Download all frame-aware nucleotide alignments (nucleotide alignments that preserve translation)
YAL015C_YOL043Cp.txt; 2 kb; Alignment for duplicated gene pair and its K. waltii ortholog (p = protein)
YAL015C_YOL043Ct.txt; 6 kb; Alignment for duplicated gene pair and its K. waltii ortholog (t = translate. Nucleotide alignment that preserves translation)
YAL017W_YOL045Wt.txt; 21 kb; Second gene pair => Frame-aware nucleotide
(...) Browse.;; Browse all alignments (nucleotide / protein/ translate)
YOR256C_YPL176Ct.txt; 12 kb; 457th gene pair => Frame-aware nucleotide
Tree_lengths_protein.txt; 25 kb; Protein-divergence branch lengths for gene pairs with S. bayanus and K. waltii orthologs
Fiveway_trees.txt; 169 kb; Protein-divergence trees for gene pairs with S. bayanus and K. waltii orthologs
Duplicated_Pairs.xls; 269 kb; Protein divergence absolute and relative rates for all pairs and their K. waltii ortholog
Nucleotide_Divergence.xls; 202 kb; Nucleotide divergence absolute and relative rates for all pairs and their K. waltii ortholog
Centromeres.xls; 17 kb; K. waltii centromere location, sequence and pairing with S. cerevisiae.
Chr6_16.jpg; 69 kb; Visualization of K. waltii centromere 1 and its pairing with Cen6:Cen16 in S. cerevisiae
Chr2_4.jpg; 117 kb; Visualization of K. waltii centromere 2 and its pairing with Cen2:Cen4 in S. cerevisiae
Chr2_4b.jpg; 62 kb; Visualization of K. waltii centromere 2 and its pairing with Cen2:Cen4 in S. cerevisiae
Chr8_11.jpg; 112 kb; Visualization of K. waltii centromere 3 and its pairing with Cen8:Cen11 in S. cerevisiae
Chr1_7.jpg; 97 kb; Visualization of K. waltii centromere 4 and its pairing with Cen1:Cen7 in S. cerevisiae
chr_10_12.jpg; 100 kb; Visualization of K. waltii centromere 5 and its pairing with Cen10:Cen12 in S. cerevisiae
Chr5_9.jpg; 123 kb; Visualization of K. waltii centromere 6 and its pairing with Cen5:Cen9 in S. cerevisiae
chr13_15.jpg; 127 kb; Visualization of K. waltii centromere 7 and its pairing with Cen13:Cen15 in S. cerevisiae
chr3_14.jpg; 111 kb; Visualization of K. waltii centromere 8 and its pairing with Cen3:Cen14 in S. cerevisiae
Telomere_hits_all.xls 947 kb Blast hits of telomeric repeat elements against K. waltii and S. cerevisiae.
Anyone you share the following link with will be able to read this content: