Play all audios:
ABSTRACT Mouse embryonic stem (ES) cells are isolated from the inner cell mass of blastocysts, and can be preserved _in vitro_ in a naive inner-cell-mass-like configuration by providing
exogenous stimulation with leukaemia inhibitory factor (LIF) and small molecule inhibition of ERK1/ERK2 and GSK3β signalling (termed 2i/LIF conditions)1,2. Hallmarks of naive pluripotency
include driving _Oct4_ (also known as _Pou5f1_) transcription by its distal enhancer, retaining a pre-inactivation X chromosome state, and global reduction in DNA methylation and in H3K27me3
repressive chromatin mark deposition on developmental regulatory gene promoters3. Upon withdrawal of 2i/LIF, naive mouse ES cells can drift towards a primed pluripotent state resembling
that of the post-implantation epiblast4. Although human ES cells share several molecular features with naive mouse ES cells5, they also share a variety of epigenetic properties with primed
murine epiblast stem cells (EpiSCs)6,7. These include predominant use of the proximal enhancer element to maintain OCT4 expression, pronounced tendency for X chromosome inactivation in most
female human ES cells, increase in DNA methylation and prominent deposition of H3K27me3 and bivalent domain acquisition on lineage regulatory genes7. The feasibility of establishing human
ground state naive pluripotency _in vitro_ with equivalent molecular and functional features to those characterized in mouse ES cells remains to be defined1. Here we establish defined
conditions that facilitate the derivation of genetically unmodified human naive pluripotent stem cells from already established primed human ES cells, from somatic cells through induced
pluripotent stem (iPS) cell reprogramming or directly from blastocysts. The novel naive pluripotent cells validated herein retain molecular characteristics and functional properties that are
highly similar to mouse naive ES cells, and distinct from conventional primed human pluripotent cells. This includes competence in the generation of cross-species chimaeric mouse embryos
that underwent organogenesis following microinjection of human naive iPS cells into mouse morulas. Collectively, our findings establish new avenues for regenerative medicine,
patient-specific iPS cell disease modelling and the study of early human development _in vitro_ and _in vivo_. Access through your institution Buy or subscribe This is a preview of
subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 51 print issues and online access $199.00 per year only
$3.90 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout
ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS GENERATION OF MOUSE–HUMAN
CHIMERIC EMBRYOS Article 02 July 2021 SELF-RENEWING HUMAN NAÏVE PLURIPOTENT STEM CELLS DEDIFFERENTIATE IN 3D CULTURE AND FORM BLASTOIDS SPONTANEOUSLY Article Open access 22 January 2024
ESTABLISHMENT OF HUMAN EXPANDED POTENTIAL STEM CELL LINES VIA PREIMPLANTATION EMBRYO CULTIVATION AND SOMATIC CELL REPROGRAMMING Article 29 April 2025 CHANGE HISTORY * _ 11 DECEMBER 2013 The
unit ‘ng ml−1’ was corrected to ’µg ml−1’, once in the Methods section and once in Extended Data Fig. 1 legend. _ REFERENCES * Hanna, J. H., Saha, K. & Jaenisch, R. Pluripotency and
cellular reprogramming: facts, hypotheses, unresolved issues. _Cell_ 143, 508–525 (2010) Article CAS Google Scholar * Ying, Q.-L. et al. The ground state of embryonic stem cell
self-renewal. _Nature_ 453, 519–523 (2008) Article ADS CAS Google Scholar * Marks, H. et al. The transcriptional and epigenomic foundations of ground state pluripotency. _Cell_ 149,
590–604 (2012) Article CAS Google Scholar * Brons, I. G. M. et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. _Nature_ 448, 191–195 (2007) Article ADS CAS
Google Scholar * Thomson, J. A. et al. Embryonic stem cell lines derived from human blastocysts. _Science_ 282, 1145–1147 (1998) Article ADS CAS Google Scholar * Chia, N.-Y. et al. A
genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. _Nature_ 468, 316–320 (2010) Article ADS CAS Google Scholar * Hanna, J. et al. Human embryonic stem
cells with biological and epigenetic characteristics similar to those of mouse ESCs. _Proc. Natl Acad. Sci. USA_ 107, 9222–9227 (2010) Article ADS CAS Google Scholar * Hanna, J. et al.
Metastable pluripotent states in NOD-mouse-derived ESCs. _Cell Stem Cell_ 4, 513–524 (2009) Article CAS Google Scholar * Greber, B. et al. Conserved and divergent roles of FGF signaling
in mouse epiblast stem cells and human embryonic stem cells. _Cell Stem Cell_ 6, 215–226 (2010) Article CAS Google Scholar * Watanabe, K. et al. A ROCK inhibitor permits survival of
dissociated human embryonic stem cells. _Nature Biotechnol._ 25, 681–686 (2007) Article CAS Google Scholar * Rajendran, G. et al. Inhibition of protein kinase C signaling maintains rat
embryonic stem cell pluripotency. _J. Biol. Chem._ 288, 24351–24362 (2013) Article CAS Google Scholar * Okamoto, I. et al. Eutherian mammals use diverse strategies to initiate
X-chromosome inactivation during development. _Nature_ 472, 370–374 (2011) Article ADS CAS Google Scholar * Mekhoubad, S. et al. Erosion of dosage compensation impacts human iPSC disease
modeling. _Cell Stem Cell_ 10, 595–609 (2012) Article CAS Google Scholar * Chappell, J., Sun, Y., Singh, A. & Dalton, S. MYC/MAX control ERK signaling and pluripotency by regulation
of dual-specificity phosphatases 2 and 7. _Genes Dev._ 27, 725–733 (2013) Article CAS Google Scholar * Betschinger, J. et al. Exit from pluripotency is gated by intracellular
redistribution of the bHLH transcription factor Tfe3. _Cell_ 153, 335–347 (2013) Article CAS Google Scholar * Rada-Iglesias, A. et al. A unique chromatin signature uncovers early
developmental enhancers in humans. _Nature_ 470, 279–283 (2011) Article ADS CAS Google Scholar * Ficz, G. et al. Fgf signaling inhibition in ESCs drives rapid genome-wide demethylation
to the epigenetic ground state of pluripotency. _Cell Stem Cell_ 13, 351–359 (2013) Article CAS Google Scholar * Rais, Y. et al. Deterministic direct reprogramming of somatic cells to
pluripotency. _Nature_ 502, 65–70 (2013) Article ADS CAS Google Scholar * Urbach, A., Bar-Nur, O., Daley, G. Q. & Benvenisty, N. Differential modeling of fragile X syndrome by human
embryonic stem cells and induced pluripotent stem cells. _Cell Stem Cell_ 6, 407–411 (2010) Article CAS Google Scholar * James, D., Noggle, S. A., Swigut, T. & Brivanlou, A. H.
Contribution of human embryonic stem cells to mouse blastocysts. _Dev. Biol._ 295, 90–102 (2006) Article CAS Google Scholar * Tachibana, M., Sparman, M., Ramsey, C., Ma, H. & Lee, H.
S. Generation of chimeric rhesus monkeys. _Cell_ 148, 285–295 (2012) Article CAS Google Scholar * Ben-Yosef, D. et al. Female sex bias in human embryonic stem cell lines. _Stem Cells
Dev._ 21, 363–372 (2012) Article CAS Google Scholar * Lengner, C. J. et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. _Cell_
141, 872–883 (2010) Article CAS Google Scholar * Takahashi, K., Mitsui, K. & Yamanaka, S. Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. _Nature_ 423,
541–545 (2003) Article ADS CAS Google Scholar * Mansour, A. A. et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. _Nature_ 488, 409–413 (2012)
Article ADS CAS Google Scholar * Massarwa, R. & Niswander, L. _In toto_ live imaging of mouse morphogenesis and new insights into neural tube closure. _Development_ 140, 226–236
(2013) Article CAS Google Scholar * Yeom, Y. I. et al. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. _Development_ 122, 881–894 (1996) CAS
PubMed Google Scholar * Pietrobono, R. et al. Quantitative analysis of DNA demethylation and transcriptional reactivation of the _FMR1_ gene in fragile X cells treated with
5-azadeoxycytidine. _Nucleic Acids Res._ 30, 3278–3285 (2002) Article CAS Google Scholar * Vassena, R. et al. Waves of early transcriptional activation and pluripotency program initiation
during human preimplantation development. _Development_ 138, 3699–3709 (2011) Article CAS Google Scholar Download references ACKNOWLEDGEMENTS J.H.H. is supported by a gift from Ilana and
Pascal Mantoux, and research grants from the European Research Council starting investigator grant (StG-2011-281906, for funding iPS cell experiments only), the Leona M. and Harry B.
Helmsley Charitable Trust, the BIRAX (Britain Israel Research and Academic Exchange Partnership), The Sir Charles Clore Research Prize, the Israel Science Foundation (Bikura, ICORE (Israeli
Centre of Research Excellence) and Regular research program), the Israel Cancer Research Foundation, the ERANET E-Rare disease program, the Benoziyo Endowment fund, Fritz Thyssen Stiftung
(used for human iPS cell experiments only), EMBO young investigator program, the Alon Foundation scholar award, a grant from E. A. and R. Drake, postdoctoral fellowships from ICRF and the
Weizmann Dean fellowship award for A.A.M. We thank the embryologists of the Racine In Vitro Fertilization Laboratory at the Lis Maternity Hospital (A. Carmon, T. Cohen and N. M. Raz) for
their skilful assistance. We thank the Weizmann Institute management for providing financial and infrastructural support. AUTHOR INFORMATION Author notes * Ohad Gafni, Leehee Weinberger,
Abed AlFatah Mansour, Yair S. Manor and Elad Chomsky: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * The Department of Molecular Genetics, Weizmann Institute of
Science, Rehovot 76100, Israel, Ohad Gafni, Leehee Weinberger, Abed AlFatah Mansour, Yair S. Manor, Elad Chomsky, Sergey Viukov, Itay Maza, Asaf Zviran, Yoach Rais, Vladislav Krupalnik,
Mirie Zerbib, Shay Geula, Inbal Caspi, Dan Schneir, Rada Massarwa, Noa Novershtern & Jacob H. Hanna * The Department of Biological Regulation, Weizmann Institute of Science, Rehovot
76100, Israel, Elad Chomsky, Zohar Shipony, Zohar Mukamel & Amos Tanay * The Department of Computer Science and Applied Mathematics, Weizmann Institute of Science, Rehovot 76100, Israel,
Elad Chomsky, Zohar Shipony, Zohar Mukamel & Amos Tanay * Wolfe PGD Stem Cell Lab, Racine IVF Unit, Lis Maternity Hospital, Tel Aviv Sourasky Medical Center, Tel-Aviv, Israel Dalit
Ben-Yosef, Yael Kalma & Tamar Shwartz * The Department of Cell and Developmental Biology, Sackler Medical School, Tel-Aviv University, Israel Dalit Ben-Yosef * The Israel National Center
for Personalized Medicine (INCPM), Weizmann Institute of Science, Rehovot 76100, Israel, Shlomit Gilad, Daniela Amann-Zalcenstein & Sima Benjamin * The Department of Immunology,
Weizmann Institute of Science, Rehovot 76100, Israel, Ido Amit Authors * Ohad Gafni View author publications You can also search for this author inPubMed Google Scholar * Leehee Weinberger
View author publications You can also search for this author inPubMed Google Scholar * Abed AlFatah Mansour View author publications You can also search for this author inPubMed Google
Scholar * Yair S. Manor View author publications You can also search for this author inPubMed Google Scholar * Elad Chomsky View author publications You can also search for this author
inPubMed Google Scholar * Dalit Ben-Yosef View author publications You can also search for this author inPubMed Google Scholar * Yael Kalma View author publications You can also search for
this author inPubMed Google Scholar * Sergey Viukov View author publications You can also search for this author inPubMed Google Scholar * Itay Maza View author publications You can also
search for this author inPubMed Google Scholar * Asaf Zviran View author publications You can also search for this author inPubMed Google Scholar * Yoach Rais View author publications You
can also search for this author inPubMed Google Scholar * Zohar Shipony View author publications You can also search for this author inPubMed Google Scholar * Zohar Mukamel View author
publications You can also search for this author inPubMed Google Scholar * Vladislav Krupalnik View author publications You can also search for this author inPubMed Google Scholar * Mirie
Zerbib View author publications You can also search for this author inPubMed Google Scholar * Shay Geula View author publications You can also search for this author inPubMed Google Scholar
* Inbal Caspi View author publications You can also search for this author inPubMed Google Scholar * Dan Schneir View author publications You can also search for this author inPubMed Google
Scholar * Tamar Shwartz View author publications You can also search for this author inPubMed Google Scholar * Shlomit Gilad View author publications You can also search for this author
inPubMed Google Scholar * Daniela Amann-Zalcenstein View author publications You can also search for this author inPubMed Google Scholar * Sima Benjamin View author publications You can also
search for this author inPubMed Google Scholar * Ido Amit View author publications You can also search for this author inPubMed Google Scholar * Amos Tanay View author publications You can
also search for this author inPubMed Google Scholar * Rada Massarwa View author publications You can also search for this author inPubMed Google Scholar * Noa Novershtern View author
publications You can also search for this author inPubMed Google Scholar * Jacob H. Hanna View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS
J.H.H. conceived the idea for this project, designed and conducted experiments, invented and optimized NHSM conditions, and wrote the manuscript with contributions from other authors. L.W.
and A.A.M. conducted tissue culture optimization and molecular biology analysis. O.G. and M.Z. conducted microinjections. O.G. and R.M. conducted embryo imaging. E.C., Z.S., Z.M. and A.T.
conducted RRBS analysis. Y.S.M. analysed gene expression data. N.N. and E.C. analyzed Chip-seq data. J.H.H., Y.K., T.S. and D.B.-Y. applied NHSM conditions on human blastocysts. O.G., I.M.,
I.A., S.Gi., D.A.-Z. and S.B. assisted in ChIP-seq experiments. S.V. and J.H.H. engineered human stem cell lines. Y.R., V.K., I.C., D.S., S.Ge. assisted and participated in iPS cell related
experimental assays. A.Z. conducted automated imaging analysis. CORRESPONDING AUTHORS Correspondence to Rada Massarwa, Noa Novershtern or Jacob H. Hanna. ETHICS DECLARATIONS COMPETING
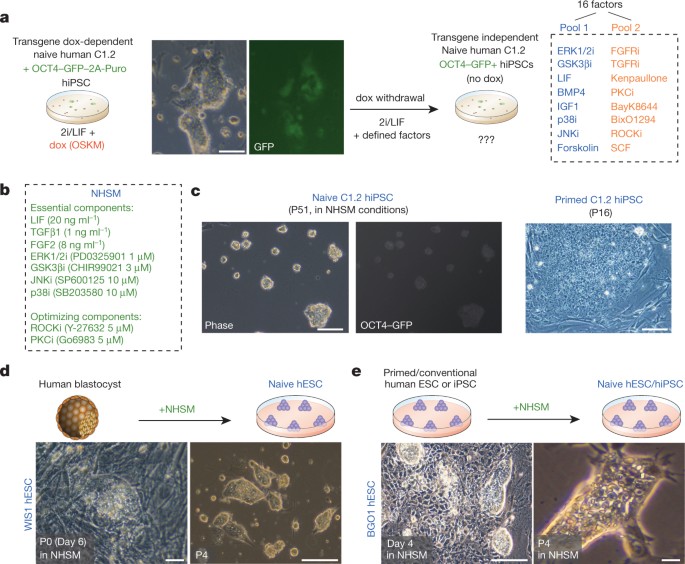
INTERESTS The authors declare no competing financial interests. EXTENDED DATA FIGURES AND TABLES EXTENDED DATA FIGURE 1 DEFINING AND OPTIMIZING CONDITIONS FOR CAPTURING TRANSGENE-INDEPENDENT
HUMAN NAIVE PLURIPOTENT CELLS. A, qRT–PCR analysis for relative expression of endogenous OCT4 pluripotency marker and SOX1 neural marker. Relative values to those measured in the primed
C1.2 iPS cell line are shown. In the presence of dox, the cells express the endogenous pluripotency OCT4 (consistent with previously described results and teratoma formation ability of these
cells7); however, SOX1 is significantly upregulated, consistent with unstable pluripotency maintenance in these conditions7. NHSM conditions optimized at the Weizmann Institute of Science
(also referred to as WIS-NHSM conditions) maintain OCT4 expression and do not acquire aberrant SOX1 upregulation. Error bars indicate s.d. (_n_ = 3). *Student’s _t_-test _P_ value < 0.05
relative to control. B, The percentage of OCT4–GFP+ colonies obtained in different combinations of indicated factors as measured at day 14. (‘−’ indicates without, ‘+’ indicates with.) Error
bars indicate s.d. (_n_ = 3). *_t_-test _P_ value, 0.01. C, Cells were expanded in the presence of FGF2/TGF-β1 and pool 1 factors, and the essential factors required for maintaining
OCT4–GFP+ cells in the absence of dox were determined by screening for loss of GFP upon withdrawal of these components (green font). *_t_-test _P_ value < 0.01. Error bars indicate s.d.
(_n_ = 4). D, OCT4–GFP+ levels for naive pluripotent cells expanded in NHSM on tissue culture plates coated with the indicated components. Naive human iPS cells can be grown without feeder
cells on Matrigel-coated plates, vitronectin-coated plates or 0.2% gelatin + 1 µg ml−1 vitronectin-coated plates. Error bars indicate s.d. (_n_ = 3). *_t_-test _P_ value < 0.01 relative
to control. E, For further optimizing naive growth conditions, 1% Albumax can be substituted by using either 15% KSR or 1% chemically defined lipid concentrate (CDLC, Invitrogen). Error bars
indicate s.d. (_n_ = 3). *_t_-test _P_ value < 0.01. This suggests the importance of lipid components in maintaining human pluripotent cells _in vitro_. F, Phase images of human naive ES
cells in the indicated conditions for 4 passages (P). G, Representative images showing embryo-derived naive human ES cells in NHSM conditions (with or without feeder cells). Notably for all
lines, naive human ES cells were transferred to primed conditions at passage 6, and cells were passaged side by side for comparative analysis as indicated throughout the study. EXTENDED
DATA FIGURE 2 VALIDATION OF HUMAN NAIVE PLURIPOTENCY. A, Karyotype analysis results indicating normal karyotypes of different naive human ES cells and human iPS cells. The passage number at
which cells were collected for karyotyping is indicated. B, The newly derived naive LIS1, LIS2, WIS1, and WIS2 human ES cell lines established from human inner cell mass in NHSM conditions
were analysed, and showed a strong uniform staining for all indicated human pluripotency markers (OCT4, NANOG, SSEA3, SSEA4, TRA1-60, TRA1-81 and SOX2). Notably, SSEA1, which is specific for
mouse (both naive and primed stem cells) and not human pluripotent cells, is not expressed on naive human ES cells. Similar results were validated for all human naive PSCs derived in this
study (data not shown). C, _In vivo_ teratoma differentiation of naive human ES cells and iPS cells. Naive pluripotent stem cell lines were expanded in NHSM conditions for the indicated
number of passages. All tested lines gave rise to well differentiated mature teratomas with cells from the three germ lineages: endoderm, ectoderm and mesoderm. P indicates passage number in
NHSM conditions at which cells were collected and injected. EXTENDED DATA FIGURE 3 GROWTH PROPERTIES OF HUMAN NAIVE PSCS. A, Population doubling time of various mouse and human pluripotent
stem cell lines. After plating each cell line in replicates, cells were collected at days 2, 4 and 6 and their growth rate was normalized by the number of cells collected and counted at day
2. Error bars represent s.d. (_n_ = 3), and _P_ values < 0.01 represent _t_-test for the average difference between primed human ES cell/human iPS cell lines and naive human ES cell/human
iPS cell lines. B, Single-cell cloning efficiency of different pluripotent stem cell lines as determined by the number of wells containing NANOG+ colonies 6 days after plating (with or
without ROCK inhibitor as indicated in the figure). *_t_-test _P_ values < 0.01 are indicated. Error bars indicate s.d. (_n_ = 3). C, Cross-species differences in ERAS protein expression
influence remaining growth-property differences between genetically unmodified human and mouse naive PSCs. Single-cell cloning efficiency of different pluripotent stem cell lines as
determined by the number of wells containing Nanog+ colonies 6 days after plating (without using ROCK inhibitor). *_t_-test _P_ value < 0.05 are indicated. Error bars indicate s.d. (_n_ =
3). These results highlight a relatively compromised single cell survival and cloning efficiency of Eras knockout naive mouse ES cells, in comparison to conventional ES cells/iPS cells.
Reconstitution of ERAS in naive human ES cells rescues their relative deficiency in single cell colony formation assay. D, Predominant utilization of distal _OCT4_ enhancer element in mouse
and human naive pluripotency. Evaluation of human _OCT4_ transcriptional regulation by the activity of distal or proximal enhancer reporter gene (by luciferase reporter assay) in the
indicated pluripotent cell lines. Baseline activity was analysed by transfection with an empty vector. Predominant utilization of distal enhancer is significantly and dramatically
upregulated in human ES cells/human iPS cells grown in naive NHSM conditions. Error bars indicate s.d. (_n_ = 3). *_t_-test _P_ value < 0.01 between the matched naive and primed samples.
E, Schematic diagram demonstrating the relative location of the evolutionary conserved distal and proximal enhancer elements in the human _OCT4_ locus. Distal enhancer (DE) and proximal
enhancer (PE) are highlighted in the human _OCT4_ locus, and were accordingly deleted in the ΔPE and ΔDE reporter constructs used in this study. Insertion site of GFP-2A-Puromycin (Puro)
resistance cassette by BAC recombineering is indicated. EXTENDED DATA FIGURE 4 EPIGENETIC PROPERTIES OF HUMAN NAIVE GROUND STATE PLURIPOTENCY. A, Fraction of cells with nuclear H3K27me3
foci. Confocal images, obtained after double immunostaining for OCT4 and H3K27me3 on naive, primed and differentiated samples, were analysed and quantified for the fraction of cells with
nuclear H3K27me3 foci that mark the inactive X allele. Average percentages of 150–200 individual cells counted per sample from independent frames are shown. Error bars indicate s.d. (_n_ =
10). *_t_-test _P_ value < 0.01 when comparing naive pluripotent cells and their derived differentiated cells. All 5 naive female pluripotent cells are predominantly devoid of H3K27me3
foci. Male lines do not exhibit H3K27me3 foci/clouds in any of the states as expected. Differentiated fibroblasts from naive cell lines uniformly acquired H3K27me3 foci. In most (3 out of 5)
primed human ES/human iPS cell lines tested, H3K27me3 foci (one per nucleus) became clearly detectable in the majority of cells consistent with XaXi configuration. Two subcloned primed cell
lines (C2 human iPS cell and WIBR2 human ES cell) showed low H3K27me3 foci abundance, consistent with XaXa or XaXe configurations known to also be acquired in human primed cell lines13. B,
qRT–PCR analysis indicates no/low expression levels of XIST in naive hPSCs, in comparison to female differentiated fibroblast cells that upregulate XIST expression. Error bars indicate s.d.
(_n_ = 3). C, Distributions of H3K9me3 RPKM (reads-per-kilobase-per-million reads) levels in chromosome X genes, measured in four different human cell lines. Boxes, 25th and 75th
percentiles; horizontal lines, median; crosses, outliers. RPKM were measured for each gene on the region starting 1 kb upstream of the TSS and ending at the TES. Lines WIBR3, C1 and LIS2 are
female and WIS2 is a male cell line. Distributions of H3K9me3 in female primed cells are significantly higher compared to their naive counterparts, whereas in male cells they are the same.
_P_ values were calculated with one-tail paired-sample _t_-test. NS, not significant. D, Bisulphite sequencing analysis of six CpG sites in single clones of an XIST promoter amplicon is
shown. Note that in female human naive cells both alleles are demethylated, whereas in primed cells one of the alleles becomes methylated. Methylation of _OCT4_ locus is shown for
comparison. E, Global H3K27me3 and H3K4me3 deposition in naive and primed pluripotent cells. Profiles of H3K27me3 and H3K4me3 chromatin modifications of all RefSeq genes in human (_n_ =
43,463) and mouse (_n_ = 30,480), naive (blue) and primed (red), represented as normalized read-density. Human profiles indicate average and s.d. (error-bars) calculated over 5 different
cell lines (C1, LIS2, WIBR3, WIBR3-MBD3mut and BGO1). Average difference between plots is indicated alongside variance and _P_ values (calculated with paired-sample _t_-test). EXTENDED DATA
FIGURE 5 DISTINCT TRANSCRIPTOME FOR HUMAN NAIVE ES CELLS/IPS CELLS. A, Genome-wide (_n_ = 12,071) transcriptional profile of the indicated primed and naive human ES cell/human iPS cell lines
measured by Affymetrix microarrays and hierarchically clustered using Pearson correlation. Heat map shows row-normalized expression levels (log2 scale) with red and green colours
representing up and downregulated genes, respectively. B, Transcriptional profile of selected pluripotency and lineage-specific marker genes, their mean expression ratio in primed human ES
cells and naive human ES cells relative to the median of all samples is presented. Values are shown in natural scale. Error bars represent s.e.m of each gene. Asterisks denote statistically
significant differentially expressed genes in which the false discovery rate was < 0.05 between the naive and primed groups of samples. C, GO (gene ontology) categories significantly
enriched for genes downregulated in naive compared to primed human cell lines, with their (–log10) FDR-corrected _P_-value. The respective mouse fold-enrichment values for the categories
that are also significant in mouse are indicated. D, Transcriptional comparison of _in vitro-_ and _in vivo_-isolated human pluripotent cells. Hierarchical clustering of the mean expression
profile of differentially expressed genes between naive and primed samples (FDR < 0.05), in the different groups (naive (this study), primed (this study), Belmonte’s primed ES cells, and
human inner cell masses (Belmonte and colleagues29)), using Spearman correlation. Note that while primed ES cell samples previously derived by Belmonte and colleagues cluster with the primed
samples derived herein, human inner cell mass samples cluster with the naive samples expanded in NHSM conditions. E, Box plot showing the median and quartiles of the distributions of the
coefficients of variance in gene expression (_σ_/_µ_) calculated for each of the 9,803 homologous genes, throughout all the samples shown in Fig. 2c. The naive distributions are
significantly smaller (one-tail _t_-test, _P_ ≈ 0) than the primed ones. EXTENDED DATA FIGURE 6 TRANSCRIPTIONAL AND MOLECULAR FEATURES OF HUMAN GROUND STATE NAIVE PLURIPOTENCY. A, Histogram
of surface expression of MHC class I using FACS analysis on the indicated naive, primed and differentiated cell lines. These results show that naive human ES cells and human iPS cells
express rudimentary or low levels of MHC class I on their cell surface, whereas primed cells upregulate MHC class I expression to intermediate levels (in comparison to differentiated 293
cells that express high levels of MHC class I). These results are identical to patterns observed in murine naive and primed cells7. B, Representative confocal immunostaining images for the
expression of E-CADHERIN and OCT4 on genetically matched naive and primed human ES cells. The samples were processed simultaneously and analysed under identical conditions. Insets represent
enlargements of boxed areas. Whereas E-CADHERIN is expressed in primed human ES cells, its expression becomes homogenously distributed and more enhanced in naive human ES cells expanded in
NHSM conditions. C, Box and whisker plots of nuclear/cytoplasmic TFE3 ratios in naive and primed mouse and human ES cells are shown. Quantitative unbiased imaging analysis for preferential
nuclear localization was conducted on randomly selected 200 cells from independent image frames per sample. Naive human ES cells showed distributions similar to those in naive mouse ES
cells, and the nuclear enrichment was lost in primed human and mouse ES cells. *_t_-test _P_ values < 1 × 10−100. D, Representative immunostaining for OCT4 and DNMT3B in human primed and
naive pluripotent cells, showing DNMT3B down regulation in the human naive ground state of pluripotency. This effect was highly dependent on efficient p38 inhibition (data not shown). E,
Representative western blot analysis for total and phosphorylated levels of the indicated proteins in genetically matched WIBR3 naive and primed human pluripotent cells (passage 25). The
results indicate blocked and reduced activity for ERK1/2, p38 and JNK in naive pluripotency. Consistent with the presence of LIF in NHSM, phosphorylated STAT3 levels accumulate in naive
human ES cells. Samples were loaded in triplicates for demonstrating consistency. Similar results were obtained when analysing naive and primed C1 human iPS cells and WIS2 human ES cells
(data not shown). SB202190 was used in NHSM conditions when p38 MAPK phosphorylation was tested. F, Naive WIS1 human ES cells expanded in NHSM conditions were expanded after omitting the
indicated factors from NHSM conditions for 72 h. qRT–PCR of lineage commitment genes is shown. The results indicate that TGF-β1 blocks the SOX1 inductive effect induced by the presence of
ERKi in the medium. Error bars indicate s.d. (_n_ = 3). *_t_-test _P_ value < 0.05. EXTENDED DATA FIGURE 7 GENOME-WIDE REDISTRIBUTION OF H3K27ME3 IN HUMAN NAIVE PLURIPOTENCY. A,
Distribution of H3K27me3 peaks in different genomic components (promoter, gene-body and intergenic region), as a function of the peak density (represented as RPKM, read per kilobase per
million reads), in human cell lines (WIBR3, C1 and BGO1). In addition to a reduction in total number of peaks in naive conditions, the peaks distribute differently, with a lower number of
peaks in promoters and gene bodies, compared to primed conditions. B, Chromatin landscape examples of 5 pluripotency genes. H3K27me3 and H3K4me3 marks are shown for naive (blue) and primed
(red) cell lines, in both human and mouse, showing high consistency between the organisms. C, Number of enhancers of class I (active transcription) and class II (poised) in naive versus
primed, human and mouse. For human, numbers correspond to common enhancers in lines WIBR3 and C1. The number of reads was normalized by down-sampling such that the number of reads in all
human samples is identical, and so is the number of reads in all mouse samples. A marked decrease in class II enhancers can be seen in the naive cells of both human and mouse. D, Expression
level distribution of genes associated with class I and class II enhancers, present in either naive cells only (blue) or in primed cells only (red). Expression level distribution of all
genes is presented as control. Boxes represent 25th and 75th quantiles, and whiskers represent minimum and maximum values that are not outliers. Analysis shows that primed state specific
class II enhancers continue to be largely repressed in both naive and primed states (despite the loss of H3K27me3 mark in naive pluripotency over their enhancer). E, Selected GO annotation
enrichment (from top 25 categories obtained) for human primed specific class II (poised) enhancer marked genes. This analysis shows that class II enhancer associated genes are highly
enriched for transcription factors (5.9 × 10−77) and developmental genes (5.5 × 10−49). EXTENDED DATA FIGURE 8 DISTINCT SIGNALLING REQUIREMENTS FOR MAINTAINING NAIVE PLURIPOTENCY ACROSS
SPECIES. A, Pluripotency maintenance by different pluripotent cell lines cultivated in various media conditions. Cross-species functionally divergent pathways are highlighted in grey. B,
Different defined growth conditions enable maintenance of mouse naive pluripotency. V6.5 mouse ES cells carrying the naive pluripotency specific ΔPE–OCT4–GFP reporter were maintained in
distinct chemically defined growth conditions as indicated in the panel labels. The analysis indicates that naive GFP+ mouse ES cells can be maintained in LIF+GSK3βi, together with Erk1/2i,
p38i or JNKi. C, Representative images of high-contribution chimaeras generated after blastocyst microinjection of the indicated lines. Agouti coat colour indicates high-level chimaera
contribution. D, Competence of different mouse naive growth conditions in tetraploid embryo complementation assay. V6.5 naive mouse ES cells were expanded in the indicated conditions for 12
passages, after which they were tested for tetraploid complementation assay to form all-ES cell animals. 135–160 mouse embryos were injected per condition tested. The analysis indicates
unrestricted developmental potential is equivalently obtained at the functional level when naive mouse ES cells are expanded with p38i, JNKi or ERK1/2i (together with supplementation of LIF
and GSK3βi in all conditions). E, Representative images of all-ES cell agouti coat coloured animals obtained. F, Different human ES cells and human iPS cell lines carrying OCT4–GFP or
ΔPE–OCT4 GFP reporter were used to evaluate different previously published conditions (as indicated in the figure panel) for their ability to maintain pluripotency and specifically naive
pluripotency (by ΔPE-Construct). Only NHSM conditions enabled maintenance of all cell lines tested while uniformly activating both OCT4–GFP and ΔPE-OCT4–GFP reporter. *Student _t_-test _P_
values < 0.01 between indicated compared samples/groups. Error bars indicate s.d. (_n_ = 3). G, LIF/STAT3 signalling dependence by naive human ES cells/iPS cells. Naive human ES cells/iPS
cells carrying a constitutively active Stat3 mutant (Stat3-CA) remain pluripotent in the absence of exogenous LIF from NHSM, based on immunostaining for pluripotency marker expression.
Error bars indicate s.d. (_n_ = 3). These results show that naive human ES cells/iPS cells described herein share defining features with rodent naive PSCs in their dependence on LIF/STAT3
signalling to remain pluripotent in NHSM conditions. EXTENDED DATA FIGURE 9 DISTINCT AND ENHANCED FUNCTIONAL PROPERTIES OF HUMAN NAIVE PLURIPOTENT CELLS. A, Targeting strategy by homologous
recombination of _COL1A_ locus in H9 ES cells. Correct targeting efficiency in different H9 human naive and primed pluripotent cell experimental replicates is shown. Correct targeting was
scored after confirmation with both 5′ and 3′ Southern blot analysis on extracted DNA from analysed clones. *Indicates significant _P_ value < 0.05. B, as in A, but for _OCT4_ locus.
*Indicates significant _P_ value < 0.05. C, NHSM conditions facilitate deterministic reprogramming of MBD3-depleted human secondary fibroblasts. MBD3wt and MBD3mut iPS cells carrying
dox-inducible OKSM transgene were labelled with constitutively expressed mCherry and targeted with an OCT4–GFP knock-in allele18. _In vitro_ differentiated fibroblasts from the latter lines
were reprogrammed as indicated in the scheme either in primed or in naive growth conditions. Secondary iPS cell reprogramming efficiencies of wild-type and MBD3-depleted (MBD3mut) cells when
reprogramming in NHSM (blue scheme) or primed (red scheme) conditions are indicated. Error bars indicate s.d. (_n_ = 3). *_t_-test _P_ value < 0.01. Deterministic up to 100% iPSC
reprogramming efficiency of primary human differentiated cells (directly obtained from human samples, rather than being differentiated _in vitro_ from pluripotent cells) remains to be
determined. D, Relative transcript expression of FMR1 and OCT4 genes in cells isolated from healthy or fragile X patients. Error bars indicate s.d. (_n_ = 3). Note that naive fragile X
patient derived iPS cells reactivate _FMR1_ gene expression. E, Methylation analysis of FMR1 promoter region on DNA samples obtained from wild-type and fragile X patient derived cells.
***Indicates student _t_-test _P_ value < 0.01. EXTENDED DATA FIGURE 10 CHIMAERISM WITH HUMAN NAIVE IPS-CELL-DERIVED CELLS FOLLOWING MOUSE MORULA MICROINJECTION. A, C1 human naive iPS
cells were targeted with constitutively CAGGS promoter driven EGFP into the human AAVS1 locus via ZFN utilization. Subsequently, cells were microinjected into E2.5 mouse early morulas, and
micro-manipulated embryos were allowed to recover and develop to blastocyst stage _ex vivo_ for an additional 24 h. Images show specific GFP+ human cell survival and integration in mouse
pre-implantation embryos. B, Naive and primed GFP+ human iPS cell survival and integration _ex vivo_, 24–36 h after microinjection into mouse morulas. *_t_-test _P_ value < 0.01. Error
bars indicate s.d. (_n_ = 3). C, Representative confocal analysis following immunostaining for GFP (green), OCT4 (red) and CDX2 (magenta) was done 24 h following mouse morula microinjections
with GFP-labelled naive human iPS cells. Note surviving GFP+ cells (white arrow) that specifically integrate and stain positive for OCT4, but not CDX2 (no co-localization between GFP and
CDX2 was observed). D, Subsequently, mouse blastocysts were allowed to develop _in vivo_, following implantation in mice, for an additional 7–8 days (as indicated), before dissection and
confocal analysis. Representative images showing robust integration of human iPS-derived cells into the neural folds of an E8.5 mouse embryo (upper panels). Notably, GFP was not detected in
the control non-injected mouse embryos (_n_ = 3, lower panels). _z_-stack interval 20 μm; 17 focal-planes total (nf, neural folds). E, Naive human iPS-cell-derived cells are integrated into
different locations at the craniofacial region of an E10.5 mouse embryo. A series of different focal planes from Fig. 4 and Supplementary Videos 1 and 2, showing multiple human
iPS-cell-derived cells integrated into different locations on the craniofacial region (arrowheads). First image in each row shows the maximum intensity projection of all _z_-stacks. The
distance between the different focal planes appears on the upper right corner of each one. fba, first branchial arch; op, otic pit; ov, optic vesicle. SUPPLEMENTARY INFORMATION SUPPLEMENTARY
INFORMATION This file contains Supplementary Discussions and additional references. (PDF 191 kb) MOUSE CROSS-SPECIES CHIMERISM WITH HUMAN NAÏVE IPSC DERIVED CELLS C1 human naïve iPSCs were
targeted with constitutively CAGGS promoter driven EGFP into the human AAVS1 locus via ZFN utilization. Subsequently cells were microinjected into E2.5 mouse morulas, and micro-manipulated
embryos were allowed to recover and develop into blastocysts in vitro. Subsequently the blastocysts were implanted in mouse uteri in vivo and allowed to develop for 7 additional days.
Embryos were dissected, stained with CellTracker and subjected to confocal imaging analysis. Video presentation of the in toto embryo imaging confocal z-sections, showing the merge of the
green (GFP) and red (CellTracker) channels of R1 inset shown in Fig. 4. (MOV 500 kb) MOUSE CROSS-SPECIES CHIMERISM WITH HUMAN NAÏVE IPSC DERIVED CELLS As, in Supplementary Video 1, but video
presentation of the in toto embryo imaging confocal z-sections, showing only the green (GFP) of R1 inset shown in Fig. 4. (MOV 113 kb) POWERPOINT SLIDES POWERPOINT SLIDE FOR FIG. 1
POWERPOINT SLIDE FOR FIG. 2 POWERPOINT SLIDE FOR FIG. 3 POWERPOINT SLIDE FOR FIG. 4 RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Gafni, O.,
Weinberger, L., Mansour, A. _et al._ Derivation of novel human ground state naive pluripotent stem cells. _Nature_ 504, 282–286 (2013). https://doi.org/10.1038/nature12745 Download citation
* Received: 19 May 2013 * Accepted: 10 October 2013 * Published: 30 October 2013 * Issue Date: 12 December 2013 * DOI: https://doi.org/10.1038/nature12745 SHARE THIS ARTICLE Anyone you share
the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer
Nature SharedIt content-sharing initiative