Play all audios:
ABSTRACT Immune regulation of cellular metabolism can be responsible for successful responses to invading pathogens. Viruses alter their hosts' cellular metabolism to facilitate
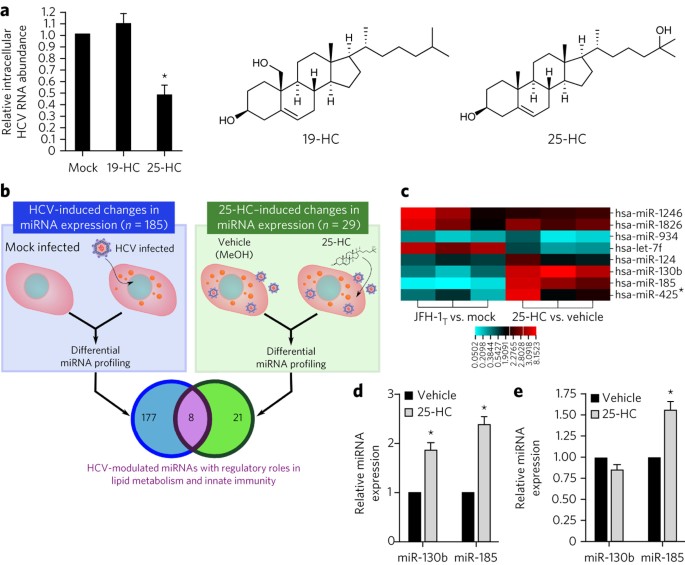
infection. Conversely, the innate antiviral responses of mammalian cells target these metabolic pathways to restrict viral propagation. We identified miR-130b and miR-185 as hepatic
microRNAs (miRNAs) whose expression is stimulated by 25-hydroxycholesterol (25-HC), an antiviral oxysterol secreted by interferon-stimulated macrophages and dendritic cells, during hepatitis
C virus (HCV) infection. However, 25-HC only directly stimulated miR-185 expression, whereas HCV regulated miR-130b expression. Independently, miR-130b and miR-185 inhibited HCV infection.
In particular, miR-185 significantly restricted host metabolic pathways crucial to the HCV life cycle. Interestingly, HCV infection decreased miR-185 and miR-130b levels to promote lipid
accumulation and counteract 25-HC's antiviral effect. Furthermore, miR-185 can inhibit other viruses through the regulation of immunometabolic pathways. These data establish these
microRNAs as a key link between innate defenses and metabolism in the liver. Access through your institution Buy or subscribe This is a preview of subscription content, access via your
institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this
article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in
* Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS _MIR-27B_ TARGETS _MAIP1_ TO MEDIATE LIPID ACCUMULATION IN
CULTURED _HUMAN_ AND _MOUSE_ HEPATIC CELLS Article Open access 24 June 2023 MIR-23A/B SUPPRESS CGAS-MEDIATED INNATE AND AUTOIMMUNITY Article Open access 25 March 2021 MACROPHAGE
MIR-4524A-5P/TBP PROMOTES Β-TRCP -TIM3 COMPLEX ACTIVATION AND TGFΒ RELEASE AND AGGRAVATES NAFLD-ASSOCIATED FIBROSIS Article Open access 19 April 2025 ACCESSION CODES PRIMARY ACCESSIONS GENE
EXPRESSION OMNIBUS * GSE73163 * GSE73164 * GSE73165 REFERENCES * Teissier, E. & Pécheur, E.I. Lipids as modulators of membrane fusion mediated by viral fusion proteins. _Eur. Biophys.
J._ 36, 887–899 (2007). Article CAS PubMed PubMed Central Google Scholar * Chukkapalli, V., Heaton, N.S. & Randall, G. Lipids at the interface of virus-host Interactions. _Curr.
Opin. Microbiol._ 15, 512–518 (2012). Article CAS PubMed PubMed Central Google Scholar * Miller, S. & Krijnse-Locker, J. Modification of intracellular membrane structures for virus
replication. _Nat. Rev. Microbiol._ 6, 363–374 (2008). Article CAS PubMed PubMed Central Google Scholar * Saka, H.A. & Valdivia, R. Emerging roles for lipid droplets in immunity and
host-pathogen interactions. _Annu. Rev. Cell Dev. Biol._ 28, 411–437 (2012). Article CAS PubMed Google Scholar * Schoggins, J.W. & Randall, G. Lipids in innate antiviral defense.
_Cell Host Microbe_ 14, 379–385 (2013). Article CAS PubMed PubMed Central Google Scholar * Blanc, M. et al. The transcription factor STAT-1 couples macrophage synthesis of
25-hydroxycholesterol to the interferon antiviral response. _Immunity_ 38, 106–118 (2013). Article CAS PubMed PubMed Central Google Scholar * Liu, S.Y. et al. Interferon-inducible
cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. _Immunity_ 38, 92–105 (2013). Article PubMed CAS Google Scholar * Pezacki, J.P. et al.
Transcriptional profiling of the effects of 25-hydroxycholesterol on human hepatocyte metabolism and the antiviral state it conveys against the hepatitis C virus. _BMC Chem. Biol._ 9, 2
(2009). Article PubMed PubMed Central CAS Google Scholar * Park, K. & Scott, A.L. Cholesterol 25-hydroxylase production by dendritic cells and macrophages is regulated by type I
interferons. _J. Leukoc. Biol._ 88, 1081–1087 (2010). Article CAS PubMed PubMed Central Google Scholar * Carthew, R.W. & Sontheimer, E.J. Origins and mechanisms of miRNAs and
siRNAs. _Cell_ 136, 642–655 (2009). Article CAS PubMed PubMed Central Google Scholar * Friedman, R.C., Farh, K.K.H., Burge, C.B. & Bartel, D.P. Most mammalian mRNAs are conserved
targets of microRNAs. _Genome Res._ 19, 92–105 (2009). Article CAS PubMed PubMed Central Google Scholar * Rottiers, V. & Näär, A.M. MicroRNAs in metabolism and metabolic disorders.
_Nat. Rev. Mol. Cell Biol._ 13, 239–250 (2012). Article CAS PubMed PubMed Central Google Scholar * Wu, J.M., Skill, N.J. & Maluccio, M.A. Evidence of aberrant lipid metabolism in
hepatitis C and hepatocellular carcinoma. _HPB (Oxford)_ 12, 625–636 (2010). Article Google Scholar * Adams, C.M. et al. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs
by different mechanisms, both involving SCAP and Insigs. _J. Biol. Chem._ 279, 52772–52780 (2004). Article CAS PubMed Google Scholar * Radhakrishnan, A., Ikeda, Y., Kwon, H.J., Brown,
M.S. & Goldstein, J.L. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. _Proc. Natl. Acad. Sci. USA_ 104,
6511–6518 (2007). Article CAS PubMed PubMed Central Google Scholar * Janowski, B.A., Willy, P.J., Devi, T.R., Falck, J.R. & Mangelsdorf, D.J. An oxysterol signalling pathway
mediated by the nuclear receptor LXRα. _Nature_ 383, 728–731 (1996). Article CAS PubMed Google Scholar * Goldstein, J.L., DeBose-Boyd, R.A. & Brown, M.S. Protein sensors for membrane
sterols. _Cell_ 124, 35–46 (2006). Article CAS PubMed Google Scholar * Su, A.I. et al. Genomic analysis of the host response to hepatitis C virus infection. _Proc. Natl. Acad. Sci. USA_
99, 15669–15674 (2002). Article CAS PubMed PubMed Central Google Scholar * Zeng, J. et al. Liver X receptors agonists impede hepatitis C virus infection in an Idol-dependent manner.
_Antiviral Res._ 95, 245–256 (2012). Article CAS PubMed Google Scholar * Russell, R.S. et al. Advantages of a single-cycle production assay to study cell culture-adaptive mutations of
hepatitis C virus. _Proc. Natl. Acad. Sci. USA_ 105, 4370–4375 (2008). Article CAS PubMed PubMed Central Google Scholar * Vlachos, I.S. et al. DIANA miRPath v.2.0: investigating the
combinatorial effect of microRNAs in pathways. _Nucleic Acids Res._ 40, W498–W504 (2012). Article CAS PubMed PubMed Central Google Scholar * Luna, J.M. et al. Hepatitis C virus RNA
functionally sequesters miR-122. _Cell_ 160, 1099–1110 (2015). Article CAS PubMed PubMed Central Google Scholar * Qadir, X.V., Han, C., Lu, D., Zhang, J. & Wu, T. miR-185 inhibits
hepatocellular carcinoma growth by targeting the DNMT1/PTEN/Akt pathway. _Am. J. Pathol._ 184, 2355–2364 (2014). Article CAS PubMed PubMed Central Google Scholar * Xiao, F. et al. A
novel function of microRNA 130a-3p in hepatic insulin sensitivity and liver steatosis. _Diabetes_ 63, 2631–2642 (2014). Article CAS PubMed Google Scholar * Steenbergen, R.H.G. et al.
Human serum leads to differentiation of human hepatoma cells, restoration of very-low-density lipoprotein secretion, and a 1000-fold increase in HCV Japanese fulminant hepatitis type 1
titers. _Hepatology_ 58, 1907–1917 (2013). Article CAS PubMed Google Scholar * Alvisi, G., Madan, V. & Bartenschlager, R. Hepatitis C virus and host cell lipids: an intimate
connection. _RNA Biol._ 8, 258–269 (2011). Article CAS PubMed Google Scholar * Pezacki, J.P., Singaravelu, R. & Lyn, R.K. Host-virus interactions during hepatitis C virus infection:
a complex and dynamic molecular biosystem. _Mol. Biosyst._ 6, 1131–1142 (2010). Article CAS PubMed Google Scholar * García-Mediavilla, M.V. et al. Liver X receptor α-mediated regulation
of lipogenesis by core and NS5A proteins contributes to HCV-induced liver steatosis and HCV replication. _Lab. Invest._ 92, 1191–1202 (2012). Article PubMed CAS Google Scholar * Waris,
G., Felmlee, D.J., Negro, F. & Siddiqui, A. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via
oxidative stress. _J. Virol._ 81, 8122–8130 (2007). Article CAS PubMed PubMed Central Google Scholar * Pezacki, J.P. et al. Chemical contrast for imaging living systems: molecular
vibrations drive CARS microscopy. _Nat. Chem. Biol._ 7, 137–145 (2011). Article CAS PubMed PubMed Central Google Scholar * Yang, M. et al. Identification of miR-185 as a regulator of de
novo cholesterol biosynthesis and low density lipoprotein uptake. _J. Lipid Res._ 55, 226–238 (2014). Article CAS PubMed PubMed Central Google Scholar * Wang, L. et al. MicroRNAs 185,
96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. _Mol. Cell. Biol._ 33, 1956–1964 (2013). Article CAS PubMed PubMed
Central Google Scholar * Pan, S., Yang, X., Jia, Y., Li, R. & Zhao, R. Microvesicle-shuttled miR-130b reduces Fat deposition in recipient primary cultured porcine adipocytes by
inhibiting PPAR-γ expression. _J. Cell. Physiol._ 229, 631–639 (2014). Article CAS PubMed Google Scholar * Hsu, P.W.C., Lin, L.Z., Hsu, S.D., Hsu, J.B.K. & Huang, H.-D. ViTa:
prediction of host microRNAs targets on viruses. _Nucleic Acids Res._ 35, D381–D385 (2007). Article CAS PubMed Google Scholar * Lyn, R.K. et al. Stearoyl-CoA desaturase inhibition blocks
formation of hepatitis C virus-induced specialized membranes. _Sci. Rep._ 4, 4549 (2014). Article PubMed PubMed Central CAS Google Scholar * Régeard, M., Trotard, M., Lepère, C.,
Gripon, P. & Le Seyec, J. Entry of pseudotyped hepatitis C virus into primary human hepatocytes depends on the scavenger class B type I receptor. _J. Viral Hepat._ 15, 865–870 (2008).
Article PubMed Google Scholar * Catanese, M.T. et al. Different requirements for scavenger receptor class B type I in hepatitis C virus cell-free versus cell-to-cell transmission. _J.
Virol._ 87, 8282–8293 (2013). Article CAS PubMed PubMed Central Google Scholar * Li, Q., Pene, V., Krishnamurthy, S., Cha, H. & Liang, T.J. Hepatitis C virus infection activates an
innate pathway involving IKK-α in lipogenesis and viral assembly. _Nat. Med._ 19, 722–729 (2013). Article CAS PubMed PubMed Central Google Scholar * Syed, G.H. et al. Hepatitis C Virus
stimulates low-density lipoprotein receptor expression to facilitate viral propagation. _J. Virol._ 88, 2519–2529 (2014). Article PubMed PubMed Central CAS Google Scholar * Monazahian,
M. et al. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. _J. Med. Virol._ 57, 223–229 (1999). Article CAS PubMed Google Scholar * Takeuchi, K. &
Reue, K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. _Am. J. Physiol. Endocrinol. Metab._ 296, E1195–E1209 (2009). Article CAS
PubMed PubMed Central Google Scholar * Mercer, D.F. et al. Hepatitis C virus replication in mice with chimeric human livers. _Nat. Med._ 7, 927–933 (2001). Article CAS PubMed Google
Scholar * Singaravelu, R. et al. Hepatitis C virus induced up-regulation of microRNA-27: A novel mechanism for hepatic steatosis. _Hepatology_ 59, 98–108 (2014). Article CAS PubMed
Google Scholar * Paul, D., Hoppe, S., Saher, G., Krijnse-Locker, J. & Bartenschlager, R. Morphological and biochemical characterization of the membranous hepatitis C virus replication
compartment. _J. Virol._ 87, 10612–10627 (2013). Article CAS PubMed PubMed Central Google Scholar * Sagan, S.M. et al. The influence of cholesterol and lipid metabolism on host cell
structure and hepatitis C virus replication. _Biochem. Cell Biol._ 84, 67–79 (2006). Article CAS PubMed Google Scholar * Li, S. et al. MicroRNA-130a inhibits HCV replication by restoring
the innate immune response. _J. Viral Hepat._ 21, 121–128 (2014). Article CAS PubMed Google Scholar * Lee, W.M. & Ahlquist, P. Membrane synthesis, specific lipid requirements, and
localized lipid composition changes associated with a positive-strand RNA Virus RNA replication protein. _J. Virol._ 77, 12819–12828 (2003). Article CAS PubMed PubMed Central Google
Scholar * Civra, A. et al. Inhibition of pathogenic non-enveloped viruses by 25-hydroxycholesterol and 27-hydroxycholesterol. _Sci. Rep._ 4, 7487 (2014). Article CAS PubMed PubMed
Central Google Scholar * Pedersen, I.M. et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. _Nature_ 449, 919–922 (2007). Article CAS PubMed PubMed Central
Google Scholar * Bassendine, M.F., Sheridan, D.A., Bridge, S.H., Felmlee, D.J. & Neely, R.D.G. Lipids and HCV. _Semin. Immunopathol._ 35, 87–100 (2013). Article CAS PubMed Google
Scholar * Blight, K.J., Kolykhalov, A.A. & Rice, C.M. Efficient initiation of HCV RNA replication in cell culture. _Science_ 290, 1972–1974 (2000). Article CAS PubMed Google Scholar
* Gong, E.Y., Fischl, W. & Bartenschlager, R. in _Antiviral Methods and Protocols_ Vol. 1030, 205–219 (Humana Press). * Kumar, A. et al. Nuclear localization of Dengue virus
nonstructural protein 5 Does Not strictly correlate with efficient viral RNA replication and inhibition of type I interferon signaling. _J. Virol._ 87, 4545–4557 (2013). Article CAS PubMed
PubMed Central Google Scholar * Stojdl, D.F. et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. _Cancer Cell_ 4, 263–275
(2003). Article CAS PubMed Google Scholar * Liu, Q.Y. et al. Identification of microRNAs involved in Alzheimer's progression using a rabbit model of the disease. _Am. J.
Neurodegener. Dis._ 3, 33–44 (2014). PubMed PubMed Central Google Scholar * Chen, J., Bardes, E.E., Aronow, B.J. & Jegga, A.G. ToppGene Suite for gene list enrichment analysis and
candidate gene prioritization. _Nucleic Acids Res._ 37, W305–W311 (2009). Article CAS PubMed PubMed Central Google Scholar * Dyer, B.W., Ferrer, F.A., Klinedinst, D.K. & Rodriguez,
R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. _Anal. Biochem._ 282, 158–161 (2000). Article CAS PubMed Google Scholar * Folch, J., Lees, M. &
Stanley, G.H.S. A simple method for the isolation and purification of total lipids from animal tissues. _J. Biol. Chem._ 226, 497–509 (1957). CAS PubMed Google Scholar * Graeve, M. &
Janssen, D. Improved separation and quantification of neutral and polar lipid classes by HPLC-ELSD using a monolithic silica phase: application to exceptional marine lipids. _J. Chromatogr.
B_ 877, 1815–1819 (2009). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We thank A. Ridsdale and the National Research Council of Canada (NRC) coherent anti-Stokes Raman
spectroscopy (CARS) facility along with Z. Jakubek and the NRC measurement science and standards (MSS) imaging facility for technical assistance. mRNA microarray profiling was performed by
the Centre for Applied Genomics, The Hospital for Sick Children, Toronto, Ontario, Canada. Lipid profiling was performed by A. Moses and the Lipid Analysis Core Service, University of
Alberta, Edmonton, Alberta, Canada. We also would like to thank E. Riklow for assistance with Dengue virus experiments. This study was supported by funding from Natural Sciences and
Engineering Research Council (NSERC) of Canada grant (298496 to J.P.P.) and Canadian Institutes of Health Research (CIHR) grants (136807, 232063 to J.P.P., R.S.R. and D.L.T.; 130365 to
K.J.R.; 28637 to T.C.H.). R.S., D.M.J., R.C. and N.G.T. would like to thank the National CIHR Research Training Program in Hepatitis C (NCRTP-HepC) for training and funding. R.S. was
supported by a Vanier Canadian Graduate scholarship. D.G.R. was supported by a CIHR graduate scholarship. D.Ö. was supported by a post-doctoral fellowship from the CIHR. A.K. was supported
by NSERC–Collaborative Research and Training Experience (CREATE) and Alberta Innovates–Health Solutions postdoctoral fellowships. T.C.H. was supported by a Tier 1 Canada Research Chair.
AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Biochemistry, Microbiology and Immunology, University of Ottawa, Ottawa, Ontario, Canada Ragunath Singaravelu, Prashanth
Srinivasan, Curtis Quan, My-Anh Nguyen, Katey J Rayner & John Paul Pezacki * Life Sciences Division, National Research Council of Canada, Ottawa, Ontario, Canada Ragunath Singaravelu,
Shifawn O'Hara, Prashanth Srinivasan, Curtis Quan, Rodney K Lyn, Dennis Özcelik, Yanouchka Rouleau & John Paul Pezacki * Immunology and Infectious Diseases, Faculty of Medicine,
Memorial University of Newfoundland, St. John's, Newfoundland, Canada Daniel M Jones, Nathan G Taylor & Rodney S Russell * Department of Medical Microbiology and Immunology,
University of Alberta and Li Ka Shing Institute of Virology, Katz Centre for Pharmacy and Health Research, Edmonton, Alberta, Canada Ran Chen, Rineke H Steenbergen & David Lorne Tyrrell
* Center for Innovative Cancer Therapeutics, Ottawa Hospital Research Institute, Ottawa, Ontario, Canada Dominic G Roy * Department of Cell Biology, University of Alberta, Edmonton, Alberta,
Canada Anil Kumar & Tom C Hobman * Department of Chemistry and Biomolecular Sciences, University of Ottawa, Ottawa, Ontario, Canada Rodney K Lyn, Dennis Özcelik & John Paul Pezacki
* University of Ottawa Heart Institute, Ottawa, Ontario, Canada My-Anh Nguyen & Katey J Rayner Authors * Ragunath Singaravelu View author publications You can also search for this author
inPubMed Google Scholar * Shifawn O'Hara View author publications You can also search for this author inPubMed Google Scholar * Daniel M Jones View author publications You can also
search for this author inPubMed Google Scholar * Ran Chen View author publications You can also search for this author inPubMed Google Scholar * Nathan G Taylor View author publications You
can also search for this author inPubMed Google Scholar * Prashanth Srinivasan View author publications You can also search for this author inPubMed Google Scholar * Curtis Quan View author
publications You can also search for this author inPubMed Google Scholar * Dominic G Roy View author publications You can also search for this author inPubMed Google Scholar * Rineke H
Steenbergen View author publications You can also search for this author inPubMed Google Scholar * Anil Kumar View author publications You can also search for this author inPubMed Google
Scholar * Rodney K Lyn View author publications You can also search for this author inPubMed Google Scholar * Dennis Özcelik View author publications You can also search for this author
inPubMed Google Scholar * Yanouchka Rouleau View author publications You can also search for this author inPubMed Google Scholar * My-Anh Nguyen View author publications You can also search
for this author inPubMed Google Scholar * Katey J Rayner View author publications You can also search for this author inPubMed Google Scholar * Tom C Hobman View author publications You can
also search for this author inPubMed Google Scholar * David Lorne Tyrrell View author publications You can also search for this author inPubMed Google Scholar * Rodney S Russell View author
publications You can also search for this author inPubMed Google Scholar * John Paul Pezacki View author publications You can also search for this author inPubMed Google Scholar
CONTRIBUTIONS R.S., K.J.R., T.C.H., D.L.T., R.S.R. and J.P.P. conceived and designed experiments. R.S., S.O'H., D.M.J., N.G.T. and R.S.R. performed cell culture and sample collection
for experiments using JFH-1T. R.S. and R.H.S. performed cell culture and sample collection for experiments using JFH-HS. R.S. and R.K.L. performed CARS microscopy experiments. R.C. and
D.L.T. performed mice experiments. M.-A.N and K.J.R. performed macrophage cell culture and sample collection. R.C. performed lipid analysis and immunofluorescence. R.S. and A.K. performed
cell culture and sample collection for experiments dealing with DENV. R.S. and D.G.R. performed cell culture and sample collection for experiments dealing with VSV. R.S., S.O'H., P.S.,
C.Q., D.Ö. and Y.R. performed all sample processing and downstream analysis. R.S., R.S.R. and J.P.P. analyzed the data. R.S. and J.P.P. wrote the manuscript. CORRESPONDING AUTHOR
Correspondence to John Paul Pezacki. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES
Supplementary Results, Supplementary Figures 1–18 and Supplementary Tables 1–6. (PDF 3099 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE
Singaravelu, R., O'Hara, S., Jones, D. _et al._ MicroRNAs regulate the immunometabolic response to viral infection in the liver. _Nat Chem Biol_ 11, 988–993 (2015).
https://doi.org/10.1038/nchembio.1940 Download citation * Received: 04 March 2015 * Accepted: 11 September 2015 * Published: 19 October 2015 * Issue Date: December 2015 * DOI:
https://doi.org/10.1038/nchembio.1940 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative