Play all audios:
ABSTRACT Whole-cell biocatalysts have proven a tractable path toward sustainable production of bulk and fine chemicals. Yet the screening of libraries of cellular designs to identify
best-performing biocatalysts is most often a low-throughput endeavor. For this reason, the development of biosensors enabling real-time monitoring of production has attracted attention. Here
we applied systematic engineering of multiple parameters to search for a general biosensor design in the budding yeast _Saccharomyces cerevisiae_ based on small-molecule binding
transcriptional activators from the prokaryote superfamily of LysR-type transcriptional regulators (LTTRs). We identified a design supporting LTTR-dependent activation of reporter gene
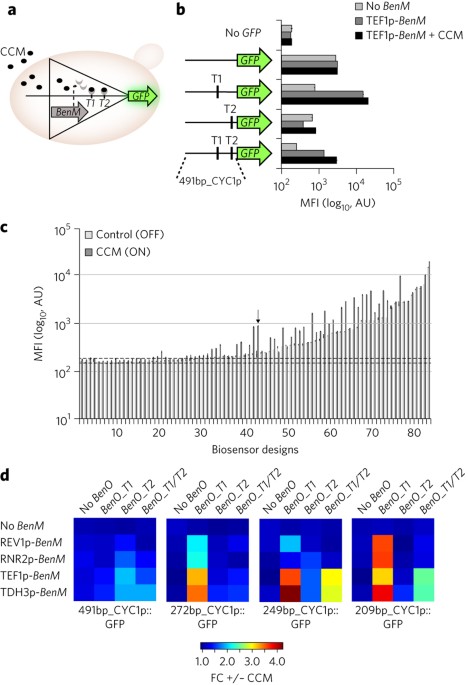
expression in the presence of cognate small-molecule inducers. As proof of principle, we applied the biosensors for _in vivo_ screening of cells producing naringenin or _cis,cis_-muconic
acid at different levels, and found that reporter gene output correlated with production. The transplantation of prokaryotic transcriptional activators into the eukaryotic chassis
illustrates the potential of a hitherto untapped biosensor resource useful for biotechnological applications. Access through your institution Buy or subscribe This is a preview of
subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $259.00 per year only
$21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout
ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS BIOSENSOR FOR BRANCHED-CHAIN
AMINO ACID METABOLISM IN YEAST AND APPLICATIONS IN ISOBUTANOL AND ISOPENTANOL PRODUCTION Article Open access 12 January 2022 ENGINEERING AND APPLICATION OF A BIOSENSOR WITH FOCUSED LIGAND
SPECIFICITY Article Open access 25 September 2020 ENGINEERING EUKARYOTE-LIKE REGULATORY CIRCUITS TO EXPAND ARTIFICIAL CONTROL MECHANISMS FOR METABOLIC ENGINEERING IN _SACCHAROMYCES
CEREVISIAE_ Article Open access 16 February 2022 ACCESSION CODES PRIMARY ACCESSIONS ARRAYEXPRESS * E-MTAB-4836 REFERENCES * Jakočiūnas, T., Jensen, M.K. & Keasling, J.D. CRISPR/Cas9
advances engineering of microbial cell factories. _Metab. Eng._ 34, 44–59 (2016). PubMed Google Scholar * Esvelt, K.M. & Wang, H.H. Genome-scale engineering for systems and synthetic
biology. _Mol. Syst. Biol._ 9, 641 (2013). PubMed PubMed Central Google Scholar * Elowitz, M.B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. _Nature_
403, 335–338 (2000). CAS PubMed Google Scholar * Wang, B., Barahona, M. & Buck, M. Amplification of small molecule-inducible gene expression via tuning of intracellular receptor
densities. _Nucleic Acids Res._ 43, 1955–1964 (2015). CAS PubMed PubMed Central Google Scholar * Farzadfard, F. & Lu, T.K. Genomically encoded analog memory with precise in vivo DNA
writing in living cell populations. _Science_ 346, 1256272 (2014). PubMed PubMed Central Google Scholar * Michener, J.K. & Smolke, C.D. High-throughput enzyme evolution in
_Saccharomyces cerevisiae_ using a synthetic RNA switch. _Metab. Eng._ 14, 306–316 (2012). CAS PubMed Google Scholar * Raman, S., Rogers, J.K., Taylor, N.D. & Church, G.M.
Evolution-guided optimization of biosynthetic pathways. _Proc. Natl. Acad. Sci. USA_ 111, 17803–17808 (2014). CAS PubMed PubMed Central Google Scholar * Choi, J.H. & Ostermeier, M.
Rational design of a fusion protein to exhibit disulfide-mediated logic gate behavior. _ACS Synth. Biol._ 4, 400–406 (2015). CAS PubMed Google Scholar * Ausländer, S., Ausländer, D.,
Müller, M., Wieland, M. & Fussenegger, M. Programmable single-cell mammalian biocomputers. _Nature_ 487, 123–127 (2012). PubMed Google Scholar * Khalil, A.S. et al. A synthetic biology
framework for programming eukaryotic transcription functions. _Cell_ 150, 647–658 (2012). CAS PubMed PubMed Central Google Scholar * Folcher, M., Xie, M., Spinnler, A. &
Fussenegger, M. Synthetic mammalian trigger-controlled bipartite transcription factors. _Nucleic Acids Res._ 41, e134 (2013). CAS PubMed PubMed Central Google Scholar * Stanton, B.C. et
al. Genomic mining of prokaryotic repressors for orthogonal logic gates. _Nat. Chem. Biol._ 10, 99–105 (2014). CAS PubMed Google Scholar * Gossen, M. & Bujard, H. Tight control of
gene expression in mammalian cells by tetracycline-responsive promoters. _Proc. Natl. Acad. Sci. USA_ 89, 5547–5551 (1992). CAS PubMed PubMed Central Google Scholar * Stanton, B.C. et
al. Systematic transfer of prokaryotic sensors and circuits to mammalian cells. _ACS Synth. Biol._ 3, 880–891 (2014). CAS PubMed PubMed Central Google Scholar * Gilbert, L.A. et al.
CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. _Cell_ 154, 442–451 (2013). CAS PubMed PubMed Central Google Scholar * Gossen, M. et al. Transcriptional
activation by tetracyclines in mammalian cells. _Science_ 268, 1766–1769 (1995). CAS PubMed Google Scholar * Teo, W.S. & Chang, M.W. Bacterial XylRs and synthetic promoters function
as genetically encoded xylose biosensors in _Saccharomyces cerevisiae_. _Biotechnol. J._ 10, 315–322 (2015). CAS PubMed Google Scholar * Lee, N., Francklyn, C. & Hamilton, E.P.
Arabinose-induced binding of AraC protein to araI2 activates the araBAD operon promoter. _Proc. Natl. Acad. Sci. USA_ 84, 8814–8818 (1987). CAS PubMed PubMed Central Google Scholar *
Shadel, G.S. & Baldwin, T.O. The Vibrio fischeri LuxR protein is capable of bidirectional stimulation of transcription and both positive and negative regulation of the luxR gene. _J.
Bacteriol._ 173, 568–574 (1991). CAS PubMed PubMed Central Google Scholar * Lee, D.J., Minchin, S.D. & Busby, S.J.W. Activating transcription in bacteria. _Annu. Rev. Microbiol._ 66,
125–152 (2012). CAS PubMed Google Scholar * Siedler, S., Stahlhut, S.G., Malla, S., Maury, J. & Neves, A.R. Novel biosensors based on flavonoid-responsive transcriptional regulators
introduced into _Escherichia coli_. _Metab. Eng._ 21, 2–8 (2014). CAS PubMed Google Scholar * Maddocks, S.E. & Oyston, P.C.F. Structure and function of the LysR-type transcriptional
regulator (LTTR) family proteins. _Microbiology_ 154, 3609–3623 (2008). CAS PubMed Google Scholar * Collier, L.S., Gaines, G.L. III & Neidle, E.L. Regulation of benzoate degradation
in Acinetobacter sp. strain ADP1 by BenM, a LysR-type transcriptional activator. _J. Bacteriol._ 180, 2493–2501 (1998). CAS PubMed PubMed Central Google Scholar * Suastegui, M. et al.
Combining Metabolic Engineering and Electrocatalysis: Application to the Production of Polyamides from Sugar. _Angew. Chem._ 128, 2414–2419 (2016). Google Scholar * Curran, K.A., Leavitt,
J.M., Karim, A.S. & Alper, H.S. Metabolic engineering of muconic acid production in _Saccharomyces cerevisiae_. _Metab. Eng._ 15, 55–66 (2013). CAS PubMed Google Scholar * Bundy,
B.M., Collier, L.S., Hoover, T.R. & Neidle, E.L. Synergistic transcriptional activation by one regulatory protein in response to two metabolites. _Proc. Natl. Acad. Sci. USA_ 99,
7693–7698 (2002). CAS PubMed PubMed Central Google Scholar * Wang, M., Li, S. & Zhao, H. Design and engineering of intracellular-metabolite-sensing/regulation gene circuits in
_Saccharomyces cerevisiae_. _Biotechnol. Bioeng._ 113, 206–215 (2016). PubMed Google Scholar * Olesen, J., Hahn, S. & Guarente, L. Yeast HAP2 and HAP3 activators both bind to the CYC1
upstream activation site, UAS2, in an interdependent manner. _Cell_ 51, 953–961 (1987). CAS PubMed Google Scholar * McIsaac, R.S., Gibney, P.A., Chandran, S.S., Benjamin, K.R. &
Botstein, D. Synthetic biology tools for programming gene expression without nutritional perturbations in _Saccharomyces cerevisiae_. _Nucleic Acids Res._ 42, e48 (2014). CAS PubMed PubMed
Central Google Scholar * Li, W.Z. & Sherman, F. Two types of TATA elements for the CYC1 gene of the yeast _Saccharomyces cerevisiae_. _Mol. Cell. Biol._ 11, 666–676 (1991). CAS
PubMed PubMed Central Google Scholar * Pfeifer, K., Arcangioli, B. & Guarente, L. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of
the CYC1 gene. _Cell_ 49, 9–18 (1987). CAS PubMed Google Scholar * Lee, M.E., Aswani, A., Han, A.S., Tomlin, C.J. & Dueber, J.E. Expression-level optimization of a multi-enzyme
pathway in the absence of a high-throughput assay. _Nucleic Acids Res._ 41, 10668–10678 (2013). CAS PubMed PubMed Central Google Scholar * Peng, H.L., Shiou, S.R. & Chang, H.Y.
Characterization of mdcR, a regulatory gene of the malonate catabolic system in Klebsiella pneumoniae. _J. Bacteriol._ 181, 2302–2306 (1999). CAS PubMed PubMed Central Google Scholar *
MacLean, A.M., MacPherson, G., Aneja, P. & Finan, T.M. Characterization of the beta-ketoadipate pathway in Sinorhizobium meliloti. _Appl. Environ. Microbiol._ 72, 5403–5413 (2006). CAS
PubMed PubMed Central Google Scholar * Laishram, R.S. & Gowrishankar, J. Environmental regulation operating at the promoter clearance step of bacterial transcription. _Genes Dev._ 21,
1258–1272 (2007). CAS PubMed PubMed Central Google Scholar * Maclean, A.M., Haerty, W., Golding, G.B. & Finan, T.M. The LysR-type PcaQ protein regulates expression of a
protocatechuate-inducible ABC-type transport system in Sinorhizobium meliloti. _Microbiology_ 157, 2522–2533 (2011). CAS PubMed Google Scholar * Chen, W.N. & Tan, K.Y. “Malonate
uptake and metabolism in _Saccharomyces cerevisiae_”. _Appl. Biochem. Biotechnol._ 171, 44–62 (2013). CAS PubMed Google Scholar * Opekarová, M. & Kubín, J. On the unidirectionality of
arginine uptake in the yeast _Saccharomyces cerevisiae_. _FEMS Microbiol. Lett._ 152, 261–267 (1997). PubMed Google Scholar * Rogers, J.K. & Church, G.M. Genetically encoded sensors
enable real-time observation of metabolite production. _Proc. Natl. Acad. Sci. USA_ 113, 2388–2393 (2016). CAS PubMed PubMed Central Google Scholar * Rikhvanov, E.G., Varakina, N.N.,
Rusaleva, T.M., Rachenko, E.I. & Voǐnikov, V.K. [The effect of sodium malonate on yeast thermotolerance]. _Mikrobiologiia_ 72, 616–620 (2003). CAS PubMed Google Scholar * Koopman, F.
et al. De novo production of the flavonoid naringenin in engineered _Saccharomyces cerevisiae_. _Microb. Cell Fact._ 11, 155 (2012). CAS PubMed PubMed Central Google Scholar *
Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. _Plant Physiol._ 126, 485–493 (2001). CAS PubMed PubMed Central
Google Scholar * Naesby, M. et al. Yeast artificial chromosomes employed for random assembly of biosynthetic pathways and production of diverse compounds in _Saccharomyces cerevisiae_.
_Microb. Cell Fact._ 8, 45 (2009). PubMed PubMed Central Google Scholar * Gupta, R.K., Patterson, S.S., Ripp, S., Simpson, M.L. & Sayler, G.S. Expression of the Photorhabdus
luminescens lux genes (luxA, B, C, D, and E) in _Saccharomyces cerevisiae_. _FEMS Yeast Res._ 4, 305–313 (2003). CAS PubMed Google Scholar * Galloway, K.E., Franco, E. & Smolke, C.D.
Dynamically reshaping signaling networks to program cell fate via genetic controllers. _Science_ 341, 1235005 (2013). PubMed PubMed Central Google Scholar * Kim, T., Folcher, M., Doaud-El
Baba, M. & Fussenegger, M. A synthetic erectile optogenetic stimulator enabling blue-light-inducible penile erection. _Angew. Chem. Int. Edn Engl._ 54, 5933–5938 (2015). CAS Google
Scholar * Zhang, H., Li, Z., Pereira, B. & Stephanopoulos, G. Engineering _E. coli_-_E. coli_ cocultures for production of muconic acid from glycerol. _Microb. Cell Fact._ 14, 134
(2015). PubMed PubMed Central Google Scholar * Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. _Nat. Protoc._ 7,
562–578 (2012). CAS PubMed PubMed Central Google Scholar * Jensen, N.B. et al. EasyClone: method for iterative chromosomal integration of multiple genes in Saccharomyces cerevisiae.
_FEMS Yeast Res._ 14, 238–248 (2014). CAS PubMed Google Scholar * Gietz, R.D. & Schiestl, R.H. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG
method. _Nat. Protoc._ 2, 38–41 (2007). CAS PubMed Google Scholar * Eckert-Boulet, N., Pedersen, M.L., Krogh, B.O. & Lisby, M. Optimization of ordered plasmid assembly by gap repair
in Saccharomyces cerevisiae. _Yeast_ 29, 323–334 (2012). CAS PubMed Google Scholar * Mikkelsen, M.D. et al. Microbial production of indolylglucosinolate through engineering of a
multi-gene pathway in a versatile yeast expression platform. _Metab. Eng._ 14, 104–111 (2012). CAS PubMed Google Scholar * Kildegaard, K.R. et al. Evolution reveals a
glutathione-dependent mechanism of 3-hydroxypropionic acid tolerance. _Metab. Eng._ 26, 57–66 (2014). CAS PubMed Google Scholar * Pettersen, E.F. et al. UCSF Chimera--a visualization
system for exploratory research and analysis. _J. Comput. Chem._ 25, 1605–1612 (2004). CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by the Novo
Nordisk Foundation and by the European Union Seventh Framework Programme (FP7-KBBE-2013-7-single-stage) under grant agreement no. 613745, Promys (M.E. & S.S.). We acknowledge A. Koza and
E. Özdemir for technical assistance. AUTHOR INFORMATION Author notes * Mette L Skjoedt and Tim Snoek: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * The Novo
Nordisk Foundation Center for Biosustainability, Technical University of Denmark, Hørsholm, Denmark Mette L Skjoedt, Tim Snoek, Kanchana R Kildegaard, Dushica Arsovska, Tobias J Goedecke,
Arun S Rajkumar, Jie Zhang, Mette Kristensen, Solvej Siedler, Irina Borodina, Michael K Jensen & Jay D Keasling * Evolva SA, Reinach, Switzerland Michael Eichenberger * Department of
Biology, Technical University Darmstadt, Darmstadt, Germany Michael Eichenberger * Evolva Biotech A/S, Copenhagen, Denmark Beata J Lehka * Department of Science and Environment, Roskilde
University, Roskilde, Denmark Beata J Lehka * Joint BioEnergy Institute, Emeryville, California, USA Jay D Keasling * Physical Biosciences Division, Lawrence Berkeley National Laboratory,
Berkeley, California, USA Jay D Keasling * Department of Chemical and Biomolecular Engineering, University of California, Berkeley, Berkeley, California, USA Jay D Keasling * Department of
Bioengineering, University of California, Berkeley, Berkeley, California, USA Jay D Keasling Authors * Mette L Skjoedt View author publications You can also search for this author inPubMed
Google Scholar * Tim Snoek View author publications You can also search for this author inPubMed Google Scholar * Kanchana R Kildegaard View author publications You can also search for this
author inPubMed Google Scholar * Dushica Arsovska View author publications You can also search for this author inPubMed Google Scholar * Michael Eichenberger View author publications You can
also search for this author inPubMed Google Scholar * Tobias J Goedecke View author publications You can also search for this author inPubMed Google Scholar * Arun S Rajkumar View author
publications You can also search for this author inPubMed Google Scholar * Jie Zhang View author publications You can also search for this author inPubMed Google Scholar * Mette Kristensen
View author publications You can also search for this author inPubMed Google Scholar * Beata J Lehka View author publications You can also search for this author inPubMed Google Scholar *
Solvej Siedler View author publications You can also search for this author inPubMed Google Scholar * Irina Borodina View author publications You can also search for this author inPubMed
Google Scholar * Michael K Jensen View author publications You can also search for this author inPubMed Google Scholar * Jay D Keasling View author publications You can also search for this
author inPubMed Google Scholar CONTRIBUTIONS M.L.S., T.S., J.D.K. and M.K.J. conceived this project. M.L.S., T.S. and M.K.J. designed all of the experiments. M.L.S., T.S. and D.A. performed
all flow cytometry analyses. M.L.S., T.S., D.A., B.J.L., J.Z., K.R.K., S.S., T.J.G. and M.E. constructed all strains and plasmids. M.K. and K.R.K. performed all analytical measurements, and
M.K.J. performed the RNA-seq experiment. M.L.S., T.S., M.K.J., I.B., A.S.R. and K.R.K. analyzed the data. M.K.J. wrote the paper. CORRESPONDING AUTHOR Correspondence to Michael K Jensen.
ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Results, Supplementary
Figures 1–7 and Supplementary Tables 1–5. (PDF 2914 kb) SUPPLEMENTARY DATASET 1 RNA-seq gene list. (XLSX 947 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Skjoedt, M., Snoek, T., Kildegaard, K. _et al._ Engineering prokaryotic transcriptional activators as metabolite biosensors in yeast. _Nat Chem Biol_ 12, 951–958 (2016).
https://doi.org/10.1038/nchembio.2177 Download citation * Received: 19 August 2015 * Accepted: 30 June 2016 * Published: 19 September 2016 * Issue Date: November 2016 * DOI:
https://doi.org/10.1038/nchembio.2177 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative