Play all audios:
ABSTRACT α-fetoprotein (AFP) is not only a widely used biomarker in hepatocellular carcinoma (HCC) surveillance, but is also clinically recognized as linked with aggressive tumour behaviour.
Here we show that deregulation of microRNA122, a liver-specific microRNA, is a cause of both AFP elevation and a more biologically aggressive phenotype in HCC. We identify CUX1, a direct
target of microRNA122, as a common central mediator of these two effects. Using liver tissues from transgenic mice in which microRNA122 is functionally silenced, an orthotopic xenograft
tumour model, and human clinical samples, we further demonstrate that a microRNA122/CUX1/microRNA214/ZBTB20 pathway regulates AFP expression. We also show that the microRNA122/CUX1/RhoA
pathway regulates the aggressive characteristics of tumours. We conclude that microRNA122 and associated signalling proteins may represent viable therapeutic targets, and that serum AFP
levels in HCC patients may be a surrogate marker for deregulated intracellular microRNA122 signalling pathways in HCC tissues. You have full access to this article via your institution.
Download PDF SIMILAR CONTENT BEING VIEWED BY OTHERS OVEREXPRESSION OF TBX3 SUPPRESSES TUMORIGENESIS IN EXPERIMENTAL AND HUMAN CHOLANGIOCARCINOMA Article Open access 22 June 2024 ANNEXINS A2
AND A5 ARE POTENTIAL EARLY BIOMARKERS OF HEPATOCARCINOGENESIS Article Open access 28 April 2023 TARGETING THE IRE1Α-XBP1S AXIS CONFERS SELECTIVE VULNERABILITY IN HEPATOCELLULAR CARCINOMA
WITH ACTIVATED WNT SIGNALING Article 28 February 2024 INTRODUCTION The incidence of hepatocellular carcinoma (HCC), the third most common cause of cancer-related mortality worldwide1, is
currently increasing2. The recent discovery of the efficacy of sorafenib, a multikinase inhibitor, as a treatment for patients with advanced HCC, has represented a major breakthrough in the
clinical management, although the survival benefit has been shown to be less than 3 months3. No other effective therapy is currently available for patients with advanced disease4. As such,
there is a continuing need to develop novel therapeutics and approaches for treatment of advanced HCC5. To develop targeted cancer therapies, we must first identify aberrantly regulated
molecular pathways specific to this cancer. Clinically, it is also important to discover useful and convenient surrogate serum biomarkers that reflect aberrations in molecular pathways due
to the molecular mechanisms of their expression, to identify the deregulated intracellular signalling pathways and to spare the patients from invasive clinical tests. Currently,
α-fetoprotein (AFP) is the most widely used serum biomarker for HCC surveillance6. Although the regulation of AFP gene expression is not fully understood, p53 (ref. 7), β-catenin8 and the
recently identified zinc-finger protein, ZBTB20 (ref. 9), have been reported to be involved. Furthermore, whereas mounting clinical evidence indicates that AFP elevation is linked to a more
aggressive tumour phenotype characterized by vascular invasion, metastasis and poor differentiation10,11, it remains to be determined whether the two phenotypes represent anything more than
coincidental epiphenomena12. MicroRNAs (miRNAs) are short, single-stranded, non-coding RNAs. Although first identified in _Caenorhabditis elegans_13, miRNAs are now known to be expressed in
most organisms, from plants to vertebrates14. Primary miRNAs, which possess stem-loop structures, are processed into mature miRNAs by Drosha and Dicer RNA polymerase III. These mature miRNAs
then associate with the RNA-induced silencing complex, and the resulting complex directly binds to the 3′-untranslated regions of target messenger RNAs to act as suppressors of translation
and gene expression. Thus, depending on the target mRNAs, miRNAs are responsible for the control of various biological functions including cell proliferation, apoptosis, differentiation,
metabolism, oncogenesis and oncogenic suppression15,16,17. MiRNA122 (miR122) is a tissue-specific miRNA that is most abundant in the liver18, wherein it is responsible for the maintenance of
fatty acid metabolism19,20 and circadian rhythms21. As shown for other tissue-specific miRNAs22, expression of miR122 has been reported to be downregulated in carcinomas, particularly in
more malignant tumours, although these results remain controversial because of conflicting reports23,24,25,26. The biological significance of the downregulation of miR122 expression in HCC
at the molecular level has not yet been fully elucidated. In the present study, we explored the role of microR122 in HCC by silencing it both in human HCC cells and in a transgenic mouse
model. Our molecular analysis enabled us to define the complex regulatory cascades underlying the clinically recognized link between raised AFP levels and a more aggressive phenotype in HCC.
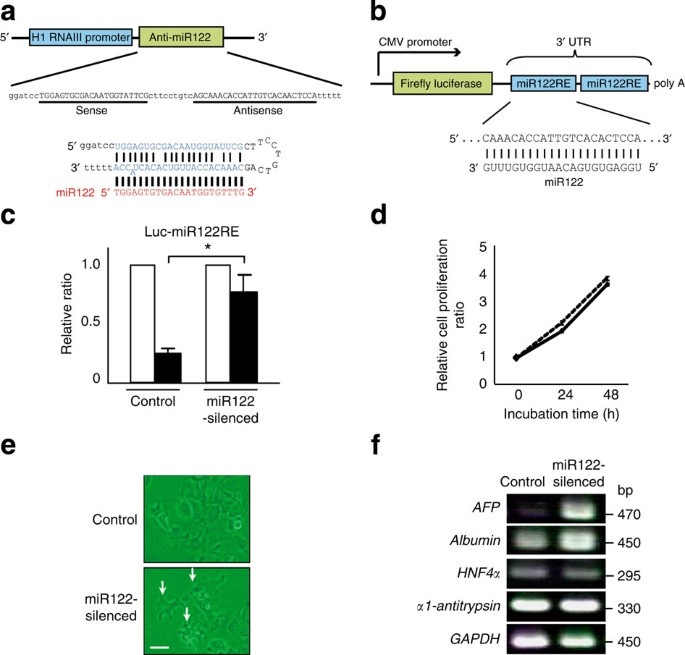
RESULTS ESTABLISHMENT OF MIR122-SILENCED HCC CELL LINES To characterize the functional consequences of miR122 downregulation in HCC cells, Huh7 and PLC/PRF/5 cells were stably transduced
with a lentivirus that expresses RNA hairpins that produce mature antisense RNA designed to silence miR122 function. These cells were selected on the basis of their relatively high levels of
miR122 expression24,27. Several mismatches were intentionally inserted into the RNA hairpin sequences to produce more stable templates for miR122 binding and sequestration and to perturb
the participation of miR122 in RNA-induced silencing complex-associated inhibition of translation (Fig. 1a). To confirm effective miR122 silencing in transduced cells, we analysed the
luciferase activity of reporters containing miR122-binding sites (the function of which is suppressed by miR122 overexpression) (Fig. 1b) in miR122-silenced and control cells. As expected,
overexpression of the miR122 precursor greatly suppressed luciferase activity in control cells (Fig. 1c). In contrast, this suppressive effect was significantly reduced in miR122-silenced
cells (Fig. 1c), indicating that miR122 was indeed functionally silenced. To characterize the biological changes that result from the loss of miR122 function, we next analysed cell
proliferation, morphology and differentiation in miR122-silenced cells. The rates of cell proliferation were comparable between control and silenced cells (Fig. 1d); however, miR122-silenced
cells exhibited a larger number of distinct pseudopodia (Fig. 1e). Next, as miR122 is specifically expressed in the liver, we hypothesized that it may have a role in hepatocyte
differentiation and, therefore, we investigated the expression of several hepatocyte markers by semi-quantitative RT–PCR. We observed an increase in AFP expression and a slight elevation of
albumin expression in miR122-silenced cells, but the expression levels of other hepatocyte markers, such as hepatocyte nuclear factor 4α (HNF4α) and α1-antitrypsin, did not change (Fig. 1f).
MIR122-SILENCED HCC CELLS EXHIBIT A MORE INVASIVE PHENOTYPE Because miR122-silenced cells exhibited an increased number of pseudopodia, we next characterized phenotypes associated with more
biologically aggressive cell characteristics. We found that actin polymerization and pseudopod formation were significantly increased in miR122-silenced cells (Fig. 2a). The increase in the
number of pseudopodia was confirmed by a quantitative pseudopodia assay (Supplementary Fig. S1). Although the expression levels of the mesenchymal marker α-smooth muscle actin were only
slightly increased, we observed a significant decrease in the expression of the epithelial marker E-cadherin in miR122-silenced cells (Fig. 2b). Furthermore, the expression of other
epithelial-to-mesenchymal transition markers such as fibronectin, N-cadherin, snail and Zeb1 was altered in miR122-silenced cells (Supplementary Fig. S1). These findings are consistent with
the notion that loss of miR122 function leads to a more malignant phenotype. We next performed scratch and invasion assays to characterize the invasive phenotype of miR122-silenced cells.
Rates of cell migration and of cell invasion were significantly increased in miR122-silenced cells (Fig. 2c,d). As the proliferation rates of control and miR122-silenced cells were similar
(Fig. 1d), altogether these results suggest that inhibition of miR122 function in HCC cells may lead to increases in malignancy-related cellular properties. To investigate the molecular
mechanisms underlying these cellular phenotypes, we assessed the activity of RhoA and Rac1,which are small GTPases that are closely associated with cell migration and invasion28. Although
Rac1 activity did not significantly change, RhoA activity significantly increased in miR122-silenced cells (Fig. 2e), suggesting that the increase in cell migration and invasion in
miR122-silenced cells may result from increased RhoA activity. AFP EXPRESSION IS INCREASED IN MIR122-SILENCED HCC CELLS As we observed an increase in AFP expression in miR122-silenced cells
(Fig. 1f), we next sought to quantify AFP concentrations in culture supernatants using an enzyme-linked immunosorbent assay (ELISA). AFP levels were approximately three times higher in the
supernatant of miR122-silenced cells as compared with control cells (Fig. 3a). Consistent with this observation, immunofluorescence staining for AFP produced a stronger cytoplasmic signal
and quantitative RT–PCR revealed a tenfold increase in AFP mRNA levels in miR122-silenced cells (Fig. 3b,c). The 3′-UTR of the AFP mRNA did not contain predicted miR122 target sequences,
based on sequence analyses performed using miRNA target search engines such as TargetScan (http://www.targetscan.org), suggesting that it is unlikely that miR122 directly regulates AFP
expression. Therefore, to characterize the mechanisms underlying increased AFP expression in miR122-silenced cells, we first assessed the stability of the AFP mRNA in miR122-silenced cells.
As expected, AFP mRNA stability was unaffected by silencing of miR122, as the amount of mRNA was comparable between control and miR122-silenced cells at 6, 12 and 24 h after inhibition of
new transcription by treatment with actinomycin D (Fig. 3d). The increase in AFP mRNA levels in the absence of changes in mRNA stability suggested that transcription of AFP was increased in
miR122-silenced cells as a result of increased AFP promoter activity. Indeed, AFP promoter activity was almost four times higher in miR122-silenced cells than in control cells, as assessed
by a reporter assay (Fig. 3e). Because AFP promoter activity is in part regulated by p53 (ref. 7), we assessed p53 activity using reporter constructs. However, no changes in p53 activity
were detected in miR122-silenced cells (Fig. 3f). As mutation of β-catenin has also been reported to be involved in upregulation of AFP expression8, we next analysed β-catenin activity in
miR122-silenced cells. Similar to p53, no change in β-catenin activity was evident in miR122-silenced cells compared with control cells (Fig. 3g). Recently, it was reported that ZBTB20 acts
as a repressor of AFP transcription9. This result led us to assess the expression of the ZBTB20 protein in miR122-silenced cells. Indeed, western blot analysis revealed that ZBTB20
expression was decreased in miR122-silenced cells (Fig. 3h). However, as ZBT20 lacks the presence of predicted miR122 target sequences based on computational searches of the 3′-UTR, it was
also unlikely that miR122 directly regulates ZBTB20 expression. These observations suggest that other indirect mechanisms may lead to decreased ZBTB20 expression in miR122-silenced cells.
CUX1 IS THE REGULATOR OF PHENOTYPES IN MIR122-SILENCED CELLS To explore the mechanisms by which miR122 regulates cell motility, invasion and AFP expression, we used computational searches to
identify potential miR122 target genes with known functions related to these processes. This analysis led to the identification of Cut homeobox 1 (CUX1, also known as CCAAT-displacement
protein/cut homeobox, CDP/Cux/Cut) through the presence of a high probability miR122 target site located in the 3′-UTR and a perfect match in the seed sequences. CUX1 is a transcription
factor that regulates multiple processes including cell cycle progression, chromosomal segregation and cell migration29,30. Consistent with the effects of miR122 silencing described above,
CUX1 was reported to modulate cell motility and invasion through the control of RhoA activity31,32,33. We observed that whereas CUX1 mRNA levels remained unchanged (Fig. 4a), there was a
significant increase in the steady-state level of the CUX1 p200 and p110 isoforms in miR122-silenced cells (Fig. 4b). To investigate the contribution of CUX1 upregulation to the increase in
AFP expression and invasive properties observed in miR122-silenced cells, we knocked down CUX1 protein expression using lentiviruses expressing CUX1 short hairpin RNAs (shRNAs) (Fig. 4c). In
the resulting double-knockdown cells, AFP protein expression in cell-culture supernatant and cell invasion were both reduced to levels similar to that of the parental Huh7 cells (Fig.
4d–f). CUX1 REPRESSES ZBTB20 EXPRESSION VIA MIR214 We next assessed whether miR122 directly targets CUX1 by constructing a luciferase reporter construct that possessed a portion of the CUX1
3′-UTR containing the putative miR122 target site (Fig. 5a). Co-transfection experiments revealed that luciferase activity was suppressed by overexpression of a miR122 precursor-expressing
plasmid (Fig. 5b). This suppressive effect was prevented by introducing two point mutations into the seed sequences of the miR122 target site (Fig. 5a,b), demonstrating that miR122 directly
targets these sequences. To confirm these effects, we generated 293T cell lines that stably expressed the miR122-precursor construct by transducing cells with miR122 precursor-expressing
lentiviruses tagged with green fluorescent protein (Supplementary Fig. S2a). As expected, the anti-miR122 construct did not affect control 293T cells, owing to the lack of miR122 expression.
However, the anti-miR122 construct greatly enhanced luciferase activity in 293T cells stably expressing the miR122-precursor, confirming that miR122 was transduced into the 293 T cells
(Supplementary Fig. S2b). Consistent with the results described above, these cells exhibited decreased expression of CUX1, particularly the p200 isoform, and also showed a modest, but
reproducible, increase in ZBTB20 expression (Fig. 5c). These results suggest that miR122 directly regulates CUX1 protein expression, which in turn may regulate ZBTB20 expression. Because
CUX1 can function as a transcriptional modulator29, we initially hypothesized that CUX1 is a direct regulator of ZBTB20 transcription. However, quantitative RT–PCR analysis revealed that
levels of the ZBTB20 mRNA were unchanged in miR122-silenced cells compared with controls (Fig. 5d). To explain the discrepancy between unchanged levels of ZBTB20 mRNA and decreases in
protein expression levels in miR122-silenced cells, we searched for miRNAs that could potentially target the ZBTB20 3′-UTR. Based on computational searches, miR214 and miR375 were identified
as candidate ZBTB20-regulatory miRNAs. Although levels of miR375 were unchanged in miR122-silenced cells (Fig. 5e), expression of miR214 was significantly increased (Fig. 5e). To assess
whether miR214 directly targeted the ZBTB20 3′-UTR, we constructed a luciferase reporter with the region of the ZBTB20 3′-UTR that contains the putative miR214 target site. Reporter assays
revealed that luciferase activity was indeed suppressed by overexpression of the miR214 precursor, suggesting that miR214 directly targets the ZBTB20 3′-UTR and suppresses its expression
(Fig. 5f). Consistent with these findings, cells that stably overexpressed the miR214 precursor exhibited decreased levels of ZBTB20 protein expression (Fig. 5g). The putative promoter
regions of miR214 contain multiple CUX1 binding sites as revealed by MATCH, a transcription factor binding site search engine (http://www.gene-regulation.com). A scanning chromatin
immunoprecipitation (ChIP) experiment, followed by real-time PCR, using a series of primer pairs, confirmed that CUX1 binds to multiple genomic sites in the miR214 promoter region (Fig. 6a).
We therefore hypothesized that CUX1 may regulate miR214 transcription. Consistent with this notion, we found that miR214 expression was decreased in CUX1 knockdown Huh7 cells (Fig. 6b). The
role of CUX1 as an activator of miR214 transcription was further verified by knocking down or overexpressing CUX1 in another cell line. Levels of miR214 decreased following the constitutive
knockdown of CUX1 with shRNA (Fig. 6c). In contrast, retroviral infection with a vector expressing p110 CUX1 led to an increase in miR214 (Fig. 6d). These findings were confirmed using
doxycycline-inducible CUX1 shRNA. As previously observed for other transcriptional targets of CUX1 (refs 30,34), levels of miR214 were reduced in the presence of doxycycline, and then
returned to levels higher than in untreated cells upon removal of the doxycycline inducer miR214 (Fig. 6e). Next, to assess the contribution of miR214 to the control of ZBTB20 expression in
miR122-silenced cells, we measured ZBTB20 expression after parallel silencing of miR214 in miR122-silenced cells. Although ZBTB20 protein expression was reduced by almost 50% by miR122
silencing, it was restored to >90% of control levels by miR214 silencing (Supplementary Fig. S3). Thus, CUX1-induced miR214 regulates, at least in part, ZBTB20 expression in
miR122-silenced cells, leading to the upregulation of AFP expression. Regulation of of CUX1 and AFP expression by miR122 was also confirmed in other HCC cell lines in which miR122 was
overexpressed or silenced. Northern blotting showed that the expression of miR122 was relatively low in Hep3B and HepG2 cells, but was relatively high in Huh1, Huh7 and PLC/PRF/5 cells
(Supplementary Fig. S4a). We therefore overexpressed the miR122 precursor in Hep3B and HepG2 cells and silenced miR122 in Huh1, Huh7 and PLC/PRF/5 cells (Supplementary Fig. S4b). CUX1
expression was respectively suppressed and enhanced by miR122 precursor overexpression and miR122 silencing (Supplementary Fig. S4c). In contrast, AFP expression was respectively enhanced
and suppressed (Supplementary Fig. S4d), confirming that AFP expression is regulated by an miR122-CUX1 pathway in multiple HCC cell lines. These results indicate that functional silencing of
miR122 leads to an increase in CUX1 protein expression, resulting in repression of ZBTB20 through an increase in miR214 expression. Repression of ZBTB20, in turn, leads to an increase in
AFP expression. Because CUX1 is a modulator of cell motility and invasion35,36,37, upregulation of this protein also enhances RhoA activity, increasing the malignant properties of cancer
cells. EXPRESSION OF CUX1-RELATED MOLECULES IN MIR122-SILENCED MICE To explore the pathway delineated above in an _in vivo_ model, we generated transgenic mice expressing antisense miR122
under the control of an H1 promoter (Fig. 7a) to inhibit the function of endogenous miR122 (ref. 38). _In situ_ hybridization analysis in these mice revealed weak miR122 staining in liver
tissue in comparison with control mice, likely due to binding of the anti-sense miR122 to endogenous miR122, which produces a double-stranded DNA and likely inhibits hybridization of the
probe (Fig. 7b). Although structural development of the liver appeared normal based on haematoxylin and eosin staining (Supplementary Fig. S5), AFP mRNA expression (Fig. 7c) and p200 and
p110 CUX1 protein expression were upregulated in the liver of anti-miR122 transgenic mice (Fig. 7d). Moreover, whereas levels of ZBTB20 mRNA were unchanged, ZBTB20 protein expression was
decreased in the liver (Fig. 7d), in agreement with _in vitro_ results demonstrating the regulation of ZBTB20 at the translational level (Fig. 5c,d). This was associated with a significant
increase in the levels of miR214 in anti-miR122 transgenic mice (Fig. 7e). Thus, results from mouse liver tissue confirm that the miR122/CUX1/miR214/ZBTB20 regulatory pathway is also
functional in an _in vivo_ model. INVASIVENESS OF MIR122-SILENCED CELLS IN XENOGRAFT MODEL Next, we transplanted control and miR122-silenced PLC/PRF/5 cells under the liver capsule of nude
mice (Fig. 7f) to determine whether miR122 silencing in HCC actually produces a more malignant phenotype _in vivo_. PLC/PRF/5 cells were chosen because of their transplantability in nude
mice39. Neither intrahepatic metastases nor vascular invasion were detected in the livers of mice transplanted with control cells at 4 weeks post-transplantation. In contrast, vascular
invasion was observed in the livers of mice transplanted with miR122-silenced HCC cells (Fig. 7g). These results suggest that miR122 silencing in HCC leads to a more aggressive phenotype.
HCC STAGING AND THE EXPRESSION OF MIR122-RELATED MOLECULES To assess the relevance of these results to human disease, we examined miR122 and AFP expression in several clinical-grade human
HCC samples. We analysed miR122 expression by _in situ_ hybridization (Fig. 8a) and AFP expression by immunohistochemistry (Fig. 8b). Both AFP expression and malignancy grading were
inversely correlated with miR122 expression levels (Fig. 8c,d). In addition, CUX1, miR214 and ZBTB20 expression was also correlated with miR122 expression, as determined using serial
sections (Supplementary Fig. S6a, b and c). These results, together with our studies in tissue culture systems and a transgenic mouse model, suggest that a reduction in the expression of
miR122 increases AFP expression via a miRNA122-CUX1-miRNA214-ZBTB20 pathway and that the development of more biologically aggressive forms of HCC occurs via a miRNA122-CUX1-RhoA pathway
(Supplementary Fig. S7). The miRNA-mediated mechanism described in this report may explain the clinically known link between increased AFP levels and more biologically aggressive cell
characteristics in HCC. DISCUSSION High AFP levels have been clinically shown to be an unfavourable prognostic factor in HCC patients40. In this study, we demonstrate that reduced expression
of miR122 in HCC cells contributes to elevated AFP expression and, subsequently, a more aggressive phenotype. These results provide a molecular framework that explains the reported link
between elevated AFP levels and a poor clinical outcome in HCC patients. Clinically, high AFP expression is correlated with more biologically aggressive properties of HCC, as patients with
high AFP levels have a significantly higher frequency of portal vein invasion and intrahepatic metastases. Additionally, these patients display significantly lower rates of recurrence-free
survival and a trend towards lower overall survival41. In the present study, we have presented several lines of evidence indicating that decreased expression of miR122 in HCC leads to the
two phenomena that are frequently observed simultaneously in the clinic: elevated expression of AFP and a more malignant biological phenotype. First, elevated AFP expression and greater
cellular invasiveness were observed in miR122-knockdown cells _in vitro_ and _in vivo_. Second, CUX1, which is linked with invasive characteristics in carcinoma cells32,36,37, was shown to
be involved in regulation of AFP expression and was identified as a direct target of miR122. Third, in human tissue samples from HCC patients, inverse correlations were observed between
miR122 expression and AFP expression, and between miR122 expression and tumour grade. These data suggest that it is unlikely that the clinical correlation between elevated AFP levels and a
more biologically aggressive phenotype in HCC is a coincidental epiphenomenon, but, instead provide a possible molecular explanation for the decrease in miR122 expression in HCC cells. A
recent study on liver development reported that liver-enriched transcription factors activate the expression of miR122, which in turn was found to promote terminal differentiation of
hepatocytes through the silencing of CUX1 (ref. 42). In the present study, we confirmed that CUX1 is a direct target of miR122 and, in contrast to the situation in normal development, we
showed that in grade 2 HCCs the decrease in miR122 is associated with higher CUX1 expression. High CUX1 expression was previously shown to inversely correlate with relapse-free and overall
survival in high-grade breast cancers36. In transgenic mice, CUX1 was reported to cause various cancer-associated disorders depending on the specific isoform and tissue type
expression34,43,44,45,46. In particular, expression of CUX1 caused organomegaly in several organs including the liver43. Hepatomegaly was associated with progression of lesions beginning
with inflammation and leading to the development of mixed cell foci, hyperplasia and even HCCs, although in this last case statistical significance was not achieved because of the small size
of the transgenic cohort47. The underlying mechanisms for the role of CUX1 in cancer is complex and is likely to involve both cell-autonomous and non-cell autonomous effects. However, from
cell-based assays it is clear that CUX1 has a role in at least three distinct processes: cell motility, cell cycle progression and chromosome segregation30,31,36,48. The knockdown of CUX1
using siRNA was shown to delay entry into S phase and to hinder cell motility and invasion31,36,48. In contrast, overexpression of p110 CUX1 was able to accelerate S phase entry and to
stimulate proliferation, migration and invasion35,48. Moreover, CUX1 was shown to promote genomic instability following cytokinesis failure30 Regulation of AFP gene expression is a complex
process mediated by a number of transcriptional activators and repressors that bind the AFP gene7,8. ZBTB20 was recently identified as a potent repressor of AFP transcription in knockout
mouse studies9. Our results demonstrate that decreased miR122 expression leads to concomitant decreases in ZBTB20 protein expression. This effect is mediated through upregulation of CUX1, as
CUX1 silencing in miR122-silenced cells was shown to lead to both recovery of ZBTB20 levels and reduced AFP expression. Furthermore, the increased expression level of ZBTB20 in CUX1
knockdown cells suggests that ZBTB20 expression is regulated by CUX1. This miR122/CUX1/miR214/ZBTB20/AFP pathway may explain the deregulated AFP expression observed in HCC cells.
Additionally, the ability of CUX1 to activate RhoA and to regulate the expression of many proteins involved in cell motility may explain the increased migration and invasiveness associated
with malignancy of HCC31,32,33,34,35,36. It should be noted that, although this analysis revealed a trend toward inverse correlation between expression of miR122 and expression of AFP, this
correlation could not be applied to all cases examined. Therefore, the possibility of additional pathways that regulate AFP expression cannot be discounted. Nonetheless, our results
demonstrate that a decrease in miR122 function is a key factor that contributes to the regulation of AFP expression in HCC. MiR122 is the most abundant miRNA in the normal adult liver,
comprising approximately 80% of all miRNAs18. The numerous reported roles of miR122 include regulation of cholesterol biosynthesis19,20, hepatitis C virus replication49 and maintenance of
the adult liver phenotype21. Specific miRNAs are often involved in the differentiation of specific cells and tissues50. As miR122 is liver-specific, we reasoned that this miRNA may have a
role in the differentiation of normal hepatocytes. In our study, transgenic mice in which miR122 was functionally silenced were found to exhibit elevated AFP levels, but did not display
abnormal morphological development in the liver (at least, not up to the age of 12 weeks), suggesting that decreased miR122 expression itself does not cause cells to become transformed.
Ongoing characterization of these mice will be required to fully determine the physiological roles of miR122 in the noncancerous liver _in vivo_. In summary, we have shown that decreased
miR122 expression in HCC is linked both to more biologically aggressive tumour behaviour and elevated AFP expression. Furthermore, both of these effects were shown to be mediated by
increased expression of CUX1, a direct target of miR122. Similar strategies could also be used to develop new therapeutics and diagnostics for other cancers in which miRNAs that regulate
both tumour characteristics and serum markers have been identified. METHODS CELL CULTURE The human HCC cell lines, Huh7, PLC/PRF/5, HepG2, Hep3B and Huh1 were obtained from the Japanese
Collection of Research Bioresources. The human embryonic kidney cell line, 293T and the human breast cancer cell line HS578T were obtained from the American Type Culture Collection . All
cells were maintained in Dulbecco's modified Eagle medium, supplemented with 10% fetal bovine serum. MOUSE EXPERIMENTS All experiments were carried out in compliance with the
regulations of the Animal Use Committee of The University of Tokyo and The Institute for Adult Disease, Asahi Life Foundation. GENERATION OF TRANSGENIC MICE IN WHICH MIR122 WAS FUNCTIONALLY
SILENCED Mice in which miR122 function was knocked down were generated using previously described protocols38,51. Briefly, a DNA fragment of 1,085 bp, containing the H1 promoter region, the
coding region for the antisense miR122 stem-loop-stem RNA precursor, and a transcriptional terminator of five thymidines, was resected from the miRZip-122 construct described above by
digestion with _Pvu_II. Proper silencing function of the resulting DNA was confirmed via transient transfection-based reporter assays that showed efficient knockdown of miR122 function.
Stable C57BL/6 embryonic stem cell lines were generated by electroporation of the linearized transgene, and the resulting cells were injected into blastocysts by the UNITECH Company.
Genotyping was performed by PCR using DNA isolated from tail snips. Four different mouse lines were maintained and the male littermates were used in experiments. CHROMATIN
IMMUNOPRECIPITATION ASSAY ChIP for CUX1 was performed as previously described52. For the scanning ChIP of the miR214 locus, realtime PCR analysis was performed using primer pairs specific
for different regions of the promoters. Templates for the PCR reactions were 0.1% total input DNA (I), nonspecific DNA from sepharose beads alone (S), or chipped chromatin. The respective
fold enrichment of the different DNA fragments are indicated relative to the DNA obtained by purification on sepharose beads without IgG (S). Enrichment was calculated using the HPRT locus
as a reference. DOXYCYCLIN-INDUCED SHRNA AGAINST CUX1 SYSTEM For conditional knockdown of CUX1 in Hs578t cells, we took advantage of the Addgene plasmid 11643. HS578T cells were infected
with pLVCT shCUX1(5,326–5,348)-tTRKRAB lentivirus as described53. At 48 h after infection, cells were split and cultured with or without doxycyclin at a final concentration of 2.5 μg ml−1.
Cells were used for experiments after 5 days of treatment. Doxycyclin was then removed from the culture media and cells were maintained for 4 days following release. CELL PROLIFERATION ASSAY
Relative cell proliferation was assessed using a Cell Counting Kit-8 (Dojindo Laboratories), as described previously54. ENZYME-LINKED IMMUNOSORBENT ASSAY AFP levels in the cell-culture
supernatant were examined using an AFP-specific ELISA kit supplied by an outsourcing company, SRL. WESTERN BLOT ANALYSIS Protein lysates were prepared from cells or mouse liver for
immunoblot analysis. Proteins were separated by SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. After blocking with 5% dry milk to decrease
nonspecific binding, membranes were probed with the appropriate primary antibodies. Primary antibodies were obtained from Abcam (ZBTB20, #ab48889, 1:500) and Santa Cruz Biotechnology (CDP,
#sc-13024, 1:1,000). CUX1 antibodies (#861, 1:1,000) were generated as described previously52. Horseradish peroxidase-conjugated secondary antibodies were used to detect primary antibodies.
Bound antibodies were detected using ECL Plus Western blotting detection reagents (GE Healthcare Life Sciences). SCRATCH ASSAY The effects of miR122 knockdown on cellular migratory function
were determined by evaluating cellular migration after scratching of a confluent monolayer of cells. Monolayers were cultured on 10 μg ml−1 fibronectin-coated dishes and were scratched using
a 200-μl pipette. Migration was analysed at the indicated time points after scratching. _IN VITRO_ INVASION ASSAY The effect of miR122 knockdown on invasive function was determined using BD
BioCoat Matrigel Invasion Chambers (Becton Dickinson) according to the manufacturer's recommended protocol. Cell invasion was induced by removing the serum in the upper chamber. The
number of invading cells was analysed after 22-h incubation. Cell numbers were counted in four randomly chosen fields at each time point. QUANTITATIVE PSEUDOPODIA ASSAY Pseudopodium
quantitation was performed using a Quantitative Pseudopodia Assay Kit (Chemicon) according to the manufacturer's instructions. Briefly, the upper chamber was coated with fibronectin and
seeded with cells in serum-free medium. Serum was added to the lower chambers. 8 h later, pseudopodia on the lower surface were stained and eluted, and the absorbances of solubilized
samples at 600 nm was measured using a microplate reader. CUX1-KNOCKDOWN LENTIVIRAL CONSTRUCT Lentiviral particles expressing CUX1 shRNA were purchased from Santa Cruz Biotechnology
(#sc-35051-V). _IN SITU_ HYBRIDIZATION TO ASSESS MIR122 AND MIR214 The expression of miR122 and miR214 in mouse liver and human HCC tissues was examined by _in situ_ hybridization55,56.
Locked nucleic acid (LNA)-scramble (negative control) and LNA-anti-miR122 and LNA-anti-miR214 probes were obtained from EXIQON. After deparaffinization, tissue sections were treated with 10
μg ml−1 proteinase K for 5 min at 37 °C and refixed with 4% paraformaldehyde, followed by acetylation with 0.25% anhydrous acetic acid in 0.1 M Tris–HCl buffer (pH 8.0). Following
pre-hybridization for 30 min at 48 °C, hybridization was performed overnight with each 20 nM LNA probe in hybridization buffer (5xSSC buffer, 50% formamide, 500 μg ml−1 tRNA, 50 μg ml−1
Cot-1 DNA). After completion of hybridization, the sections were washed with 0.1×SSC buffer for 10 min at 52 °C three times and blocked with DIG blocking buffer (Roche Diagnostics) for 30
min. Sections were then probed with anti-DIG (1:500; Roche Diagnostics) for 1 h at room temperature. Detection was performed by incubation in NBT/BCIP buffer (Promega) overnight. Nuclei were
stained with Nuclear FastRed (Sigma-Aldrich). IMMUNOHISTOCHEMISTRY Tissue arrays containing HCC tissues were purchased from US Biomax. To determine the correlations between AFP, ZBTB20,
CUX1, miR122 and miR214 expression and HCC differentiation grade, slides carrying consecutive sections were obtained. Slides were baked at 65 °C for 1 h and deparaffinized. Endogenous
peroxidase activity was blocked by incubation in 3% hydrogen peroxide buffer for 30 min. Antigen retrieval was performed by incubating the slides at 89 °C in 10 mM sodium citrate buffer (pH
6.0) for 30 min. To minimize nonspecific background staining, slides were blocked in 5% normal goat serum (Dako) for 10 min at room temperature. Tissues were labelled overnight at 4 °C with
primary antibodies raised against AFP (Dako, #N1501, 1:100), CUX1 (#sc-13024, 1:100) and ZBTB20 (HPA016815, Sigma-Aldrich, 1:100). Slides were then incubated with anti-rabbit horseradish
peroxidase-conjugated secondary antibodies (Nichirei Bioscience) for 1 h. Primary antibody binding was visualized by incubation in 3,3′-diaminobenzidine in buffered substrate (Nichirei
Bioscience) for 5 min. The slides were counterstained with haematoxylin, dehydrated with ethanol, and mounted with Clarion mounting medium (Biomeda). GTP-BINDING RHOA AND RAC1
IMMUNOPRECIPITATION ASSAY The amount of RhoA activity was examined using an Active Rho Pull-down and Detection Kit (Thermo Fisher Scientific) according to the manufacturer's recommended
protocol. The amount of GTP-bound RhoA protein (the active form of RhoA) was detected by Western blotting with the provided anti-RhoA antibody (1:100). Rac1 activity was similarly
determined by using PAK-GST Protein Beads (#PAK02, Cytoskeleton) for pulldowns and anti-Rac1 antibodies (1:100) for subsequent Western blotting (#89856D, Thermo Fisher Scientific).
ORTHOTOPIC XENOGRAFT TUMOUR MODEL OF HCC Male BALB/c (nu/nu) nude mice were purchased from CREA Japan (Tokyo, Japan). The transplantation of tumour cells into mouse livers was performed
using previously reported methods57,58. Briefly, 2×106 control or miR122-silenced PLC/PRF5 cells were suspended in 30 μl of PBS containing 1% Matrigel (Becton Dickinson). After anaesthesia,
the liver was exposed through a surgical incision. Cells were slowly injected under the capsule of left lobe of the liver using a 28-gauge needle. When successful, a transparent bleb of
cells could be seen through the liver capsule. After injection, light pressure was applied to the injection site with sterile gauze for 2 min to prevent bleeding and tumour cell leakage. The
abdomen was then closed with sutures. Transplantation was successful in a total of 12 mice (6/group). At 4 weeks post-transplantation, liver tissues were collected, serially sectioned, and
stained with haematoxylin and eosin. STATISTICAL ANALYSIS Statistically significant differences between groups were determined using Student's _t_-test when variances were equal. When
variances were unequal, Welch's _t_-test was used instead. _P_-values less than 0.05 were considered statistically significant. Plasmid and stable cell line construction, reporter
assays, RT–PCR, northern blotting and immunocytochemistry are described in the Supplementary Methods. All primer information is provided in Supplementary Table S1. ADDITIONAL INFORMATION HOW
TO CITE THIS ARTICLE: Kojima, K. _et al_. MiRNA122 is a key regulator of α-fetoprotein expression and influences the aggressiveness of hepatocellular carcinoma. _Nat. Commun._ 2:338 doi:
10.1038/ncomms1345 (2011). CHANGE HISTORY * _ 25 SEPTEMBER 2012 A correction has been published and is appended to both the HTML and PDF versions of this paper. The error has not been fixed
in the paper. _ REFERENCES * Parkin, D., Bray, F., Ferlay, J. & Pisani, P. Global cancer statistics, 2002. _CA Cancer J. Clin._ 55, 74–108 (2005). Article PubMed Google Scholar *
El-Serag, H. Epidemiology of hepatocellular carcinoma in USA. _Hepatol. Res._ 37, S88–94 (2007). Article PubMed Google Scholar * Llovet, J. et al. Sorafenib in advanced hepatocellular
carcinoma. _N. Engl. J. Med._ 359, 378–390 (2008). Article CAS PubMed Google Scholar * Greten, T. et al. _S_urvival rate in patients with hepatocellular carcinoma: a retrospective
analysis of 389 patients. _Br. J. Cancer_ 92, 1862–1868 (2005). Article CAS PubMed PubMed Central Google Scholar * Greten, T., Korangy, F., Manns, M. & Malek, N. Molecular therapy
for the treatment of hepatocellular carcinoma. _Br. J. Cancer_ 100, 19–23 (2009). Article CAS PubMed Google Scholar * Di Bisceglie, A. Issues in screening and surveillance for
hepatocellular carcinoma. _Gastroenterology_ 127, S104–S107 (2004). Article PubMed Google Scholar * Ogden, S. et al. _p53_ targets chromatin structure alteration to repress
alpha-fetoprotein gene expression. _J. Biol. Chem._ 276, 42057–42062 (2001). Article CAS PubMed Google Scholar * Peng, S. et al. High alpha-fetoprotein level correlates with high stage,
early recurrence and poor prognosis of hepatocellular carcinoma: significance of hepatitis virus infection, age, p53 and beta-catenin mutations. _Int. J. Cancer._ 112, 44–50 (2004). Article
CAS PubMed Google Scholar * Xie, Z. et al. Zinc finger protein ZBTB20 is a key repressor of alpha-fetoprotein gene transcription in liver. _Proc. Natl Acad. Sci. USA_ 105, 10859–10864
(2008). Article ADS CAS PubMed PubMed Central Google Scholar * Oishi, K. et al. Clinicopathologic features of poorly differentiated hepatocellular carcinoma. _J. Surg. Oncol._ 95,
311–316 (2007). Article PubMed Google Scholar * Yamamoto, K. et al. AFP, AFP-L3, DCP, and GP73 as markers for monitoring treatment response and recurrence and as surrogate markers of
clinicopathological variables of HCC. _J. Gastroenterol._ 45, 1272–1282 (2010). Article ADS CAS PubMed Google Scholar * Matsumoto, Y. et al. Clinical classification of hepatoma in Japan
according to serial changes in serum alpha-fetoprotein levels. _Cancer_ 49, 354–360 (1982). Article CAS PubMed Google Scholar * Lee, R., Feinbaum, R. & Ambros, V. The _C. elegans_
heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. _Cell_ 75, 843–854 (1993). Article CAS PubMed Google Scholar * Carrington, J. & Ambros, V. Role
of microRNAs in plant and animal development. _Science_ 301, 336–338 (2003). Article ADS CAS PubMed Google Scholar * Bartel, D. MicroRNAs: genomics, biogenesis, mechanism, and function.
_Cell_ 116, 281–297 (2004). Article CAS PubMed Google Scholar * Ambros, V. The functions of animal microRNAs. _Nature_ 431, 350–355 (2004). Article ADS CAS PubMed Google Scholar *
Lu, J. et al. MicroRNA expression profiles classify human cancers. _Nature_ 435, 834–838 (2005). Article ADS CAS PubMed Google Scholar * Landgraf, P. et al. A mammalian microRNA
expression atlas based on small RNA library sequencing. _Cell_ 129, 1401–1414 (2007). Article CAS PubMed PubMed Central Google Scholar * Krützfeldt, J. et al. Silencing of microRNAs _in
vivo_ with 'antagomirs'. _Nature_ 438, 685–689 (2005). Article ADS PubMed Google Scholar * Esau, C. et al. miR-122 regulation of lipid metabolism revealed by _in vivo_
antisense targeting. _Cell Metab._ 3, 87–98 (2006). Article CAS PubMed Google Scholar * Gatfield, D. et al. Integration of microRNA miR-122 in hepatic circadian gene expression. _Genes
Dev._ 23, 1313–1326 (2009). Article CAS PubMed PubMed Central Google Scholar * Yan, D. et al. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. _J. Biol. Chem._
284, 29596–29604 (2009). Article CAS PubMed PubMed Central Google Scholar * Kutay, H. et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. _J. Cell.
Biochem._ 99, 671–678 (2006). Article CAS PubMed PubMed Central Google Scholar * Coulouarn, C., Factor, V., Andersen, J., Durkin, M. & Thorgeirsson, S. Loss of miR-122 expression in
liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. _Oncogene_ 28, 3526–3536 (2009). Article CAS PubMed PubMed Central Google Scholar *
Tsai, W. et al. MicroRNA-122 a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. _Hepatology_ 49, 1571–1582 (2009). Article CAS PubMed Google
Scholar * Varnholt, H. et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. _Hepatology_ 47, 1223–1232 (2008). Article CAS PubMed Google
Scholar * Wong, Q. et al. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. _Gastroenterology_ 135, 257–269 (2008). Article CAS
PubMed Google Scholar * Sahai, E. & Marshall, C. RHO-GTPases and cancer. _Nat. Rev. Cancer_ 2, 133–142 (2002). Article PubMed Google Scholar * Sansregret, L. & Nepveu, A. The
multiple roles of CUX1: insights from mouse models and cell-based assays. _Gene_ 412, 84–94 (2008). Article CAS PubMed Google Scholar * Sansregret, L. et al. Cut homeobox 1 causes
chromosomal instability by promoting bipolar division after cytokinesis failure. _Proc. Natl Acad. Sci. USA_ 108, 1949–1954 (2011). Article ADS CAS PubMed PubMed Central Google Scholar
* Kedinger, V. & Nepveu1, A. The roles of CUX1 homeodomain proteins in the establishment of a transcriptional program required for cell migration and invasion. _Cell Adh. Migr._ 4,
348–352 (2010). Article PubMed PubMed Central Google Scholar * Michl, P., Knobel, B. & Downward, J. CUTL1 is phosphorylated by protein kinase A, modulating its effects on cell
proliferation and motility. _J. Biol. Chem._ 281, 15138–15144 (2006). Article CAS PubMed Google Scholar * Seguin, L. et al. CUX1 and E2F1 regulate coordinated expression of the mitotic
complex genes Ect2, MgcRacGAP, and MKLP1 in S phase. _Mol. Cell Biol._ 29, 570–581 (2009). Article CAS PubMed Google Scholar * Kedinger, V. et al. p110 CUX1 homeodomain protein
stimulates cell migration and invasion in part through a regulatory cascade culminating in the repression of E-cadherin and occludin. _J. Biol. Chem._ 284, 27701–27711 (2009). Article CAS
PubMed PubMed Central Google Scholar * Michl, P. et al. CUTL1 is a target of TGF(beta) signaling that enhances cancer cell motility and invasiveness. _Cancer Cell_ 7, 521–532 (2005).
Article CAS PubMed Google Scholar * Aleksic, T. et al. CUTL1 promotes tumor cell migration by decreasing proteasome-mediated Src degradation. _Oncogene_ 26, 5939–5949 (2007). Article
CAS PubMed Google Scholar * Kunath, T. et al. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. _Nat. Biotechnol._ 21, 559–561 (2003). Article
ADS CAS PubMed Google Scholar * Shouval, D. et al. Tumorigenicity in nude mice of a human hepatoma cell line containing hepatitis B virus DNA. _Cancer Res._ 41, 1342–1350 (1981). CAS
PubMed Google Scholar * Nomura, F., Ohnishi, K. & Tanabe, Y. Clinical features and prognosis of hepatocellular carcinoma with reference to serum alpha-fetoprotein levels. Analysis of
606 patients. _Cancer_ 64, 1700–1707 (1989). Article CAS PubMed Google Scholar * Johnson, P., Melia, W., Palmer, M., Portmann, B. & Williams, R. Relationship between serum
alpha-foetoprotein, cirrhosis and survival in hepatocellular carcinoma. _Br. J. Cancer_ 44, 502–505 (1981). Article CAS PubMed PubMed Central Google Scholar * Xu, H. et al.
Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. _Hepatology_ 52, 1431–1442 (2010). Article CAS PubMed Google Scholar * Ledford, A.
W. et al. Deregulated expression of the homeobox gene Cux-1 in transgenic mice results in downregulation of p27(kip1) expression during nephrogenesis, glomerular abnormalities, and
multiorgan hyperplasia. _Dev. Biol._ 245, 157–171 (2002). Article MathSciNet CAS PubMed PubMed Central Google Scholar * Brantley, J. G., Sharma, M., Alcalay, N. I. & Heuvel, G. B.
Cux-1 transgenic mice develop glomerulosclerosis and interstitial fibrosis. _Kidney Int._ 63, 1240–1248 (2003). Article PubMed Google Scholar * Cadieux, C. et al. Mouse mammary tumor
virus p75 and p110 CUX1 transgenic mice develop mammary tumors of various histologic types. _Cancer Res._ 69, 7188–7197 (2009). Article CAS PubMed Google Scholar * Cadieux, C. et al.
Polycystic kidneys caused by sustained expression of Cux1 isoform p75. _J. Biol. Chem._ 283, 13817–13824 (2008). Article CAS PubMed Google Scholar * Cadieux, C. et al. Transgenic mice
expressing the p75 CCAAT-displacement protein/Cut homeobox isoform develop a myeloproliferative disease-like myeloid leukemia. _Cancer Res._ 66, 9492–9501 (2006). Article CAS PubMed
Google Scholar * Vanden Heuvel, G. B. et al. Hepatomegaly in transgenic mice expressing the homeobox gene Cux-1. _Mol. Carcinog._ 43, 18–30 (2005). Article CAS PubMed PubMed Central
Google Scholar * Sansregret, L. et al. The p110 isoform of the CDP/Cux transcription factor accelerates entry into S phase. _Mol. Cell Biol._ 26, 2441–2455 (2006). Article CAS PubMed
PubMed Central Google Scholar * Jopling, C., Yi, M., Lancaster, A., Lemon, S. & Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. _Science_ 309,
1577–1581 (2005). Article ADS CAS PubMed Google Scholar * Taulli, R. et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by
promoting myogenic differentiation. _J. Clin. Invest._ 119, 2366–2378 (2009). CAS PubMed PubMed Central Google Scholar * Zhou, Y. et al. Chimeric mouse tumor models reveal differences in
pathway activation between ERBB family- and KRAS-dependent lung adenocarcinomas. _Nat. Biotechnol._ 28, 71–78 (2010). Article CAS PubMed Google Scholar * Harada, R. et al. Genome-wide
location analysis and expression studies reveal a role for p110 CUX1 in the activation of DNA replication genes. _Nucleic Acids Res._ 36, 189–202 (2008). Article CAS PubMed Google Scholar
* Szulc, J., Wiznerowicz, M., Sauvain, M. O., Trono, D. & Aebischer, P. A versatile tool for conditional gene expression and knockdown. _Nat. Methods_ 3, 109–116 (2006). Article CAS
PubMed Google Scholar * Maeda, S. et al. Ikappa B kinasebeta/nuclear factor-kappaB activation controls the development of liver metastasis by way of interleukin-6 expression. _Hepatology_
50, 1851–1860 (2009). Article CAS PubMed Google Scholar * Elmén, J. et al. LNA-mediated microRNA silencing in non-human primates. _Nature_ 452, 896–899 (2008). Article ADS PubMed
Google Scholar * Bai, S. et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. _J. Biol. Chem._ 284, 32015–32027
(2009). Article CAS PubMed PubMed Central Google Scholar * Yao, X. et al. A novel orthotopic tumor model to study growth factors and oncogenes in hepatocarcinogenesis. _Clin. Cancer
Res._ 9, 2719–2726 (2003). CAS PubMed Google Scholar * Kim, M. et al. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. _Nat. Protoc._
4, 1670–1680 (2009). Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by Grants-in-Aid from the Ministry of Education,
Culture, Sports, Science and Technology, Japan (#22390058, #22590718, #17016016 and #20390204) (to M. Otsuka, Y. Kondo, M. Omata and K. Koike), by Health Sciences Research Grants of The
Ministry of Health, Labour and Welfare of Japan (Research on Hepatitis) (to K. Koike), by grants from the Takeda Science Foundation, Astellas Foundation for Research on Metabolic Disorders,
Senri Life Science Foundation, the Foundation for Promotion of Cancer Research and the Mochida Memorial Foundation for Medical and Pharmaceutical Research (to M. Otsuka), and by the grant
019389 from the Canadian Cancer Society (to A.N.). AUTHOR INFORMATION Author notes * Kentaro Kojima, Akemi Takata and Charles Vadnais: These authors contributed equally to this work. AUTHORS
AND AFFILIATIONS * Department of Gastroenterology, Graduate School of Medicine, The University of Tokyo, Tokyo, 113-8655, Japan Kentaro Kojima, Akemi Takata, Motoyuki Otsuka, Takeshi
Yoshikawa, Yuji Kondo, Takahiro Kishikawa, Haruhiko Yoshida, Masao Omata & Kazuhiko Koike * Goodman Cancer Center and Departments of Oncology, Biochemistry and Medicine, McGill
University, Montreal, H3A 1A3, Quebec, Canada Charles Vadnais & Alain Nepveu * Division of Gastroenterology, The Institute for Adult Diseases, Asahi Life Foundation, Tokyo, 100-0005,
Japan Masao Akanuma * Department of Immunology and Microbial Science, The Scripps Research Institute, La Jolla, 92037, California, USA Young Jun Kang * Unit of Disease Control Genome
Medicine, The Institute of Medical Science, The University of Tokyo, Tokyo, 108-8639, Japan Naoya Kato * Department of Pathophysiology, Second Military Medical University, Shanghai, 200433,
China Zhifang Xie & Weiping J. Zhang Authors * Kentaro Kojima View author publications You can also search for this author inPubMed Google Scholar * Akemi Takata View author publications
You can also search for this author inPubMed Google Scholar * Charles Vadnais View author publications You can also search for this author inPubMed Google Scholar * Motoyuki Otsuka View
author publications You can also search for this author inPubMed Google Scholar * Takeshi Yoshikawa View author publications You can also search for this author inPubMed Google Scholar *
Masao Akanuma View author publications You can also search for this author inPubMed Google Scholar * Yuji Kondo View author publications You can also search for this author inPubMed Google
Scholar * Young Jun Kang View author publications You can also search for this author inPubMed Google Scholar * Takahiro Kishikawa View author publications You can also search for this
author inPubMed Google Scholar * Naoya Kato View author publications You can also search for this author inPubMed Google Scholar * Zhifang Xie View author publications You can also search
for this author inPubMed Google Scholar * Weiping J. Zhang View author publications You can also search for this author inPubMed Google Scholar * Haruhiko Yoshida View author publications
You can also search for this author inPubMed Google Scholar * Masao Omata View author publications You can also search for this author inPubMed Google Scholar * Alain Nepveu View author
publications You can also search for this author inPubMed Google Scholar * Kazuhiko Koike View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS
K. Kojima, M. Otsuka and A.N. planned the research and wrote the paper. K. Kojima, A.T., C.V., T.Y., Y. Kondo, Y. Kang and Z.X. performed the majority of the experiments. M.A., N.K., W.Z.
and A.N. contributed materials. T.K. and H.Y. supported several experiments. M. Omata and K. Koike supervised the entire project. CORRESPONDING AUTHOR Correspondence to Motoyuki Otsuka.
ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Figures S1 – S7, Supplementary
Table S1, Supplementary Methods and Supplementary Reference. (PDF 562 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Kojima, K., Takata, A.,
Vadnais, C. _et al._ MicroRNA122 is a key regulator of α-fetoprotein expression and influences the aggressiveness of hepatocellular carcinoma. _Nat Commun_ 2, 338 (2011).
https://doi.org/10.1038/ncomms1345 Download citation * Received: 30 March 2011 * Accepted: 11 May 2011 * Published: 07 June 2011 * DOI: https://doi.org/10.1038/ncomms1345 SHARE THIS ARTICLE
Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided
by the Springer Nature SharedIt content-sharing initiative