Play all audios:
ABSTRACT Hematopoietic stem cells give rise to progeny that either self-renew in an undifferentiated state or lose self-renewal capabilities and commit to lymphoid or myeloid lineages. Here
we evaluated whether hematopoietic stem cell self-renewal is affected by the Notch pathway. Notch signaling controls cell fate choices in both invertebrates and vertebrates1,2,3,4,5,6,7 by
inhibiting certain differentiation pathways, thereby permitting cells to either differentiate along an alternative pathway or to self-renew1. Notch receptors are present in hematopoietic
precursors and Notch signaling enhances the _in vitro_ generation of human and mouse hematopoietic precursors8,9,10,11,12,13,14,15, determines T- or B-cell lineage specification from a
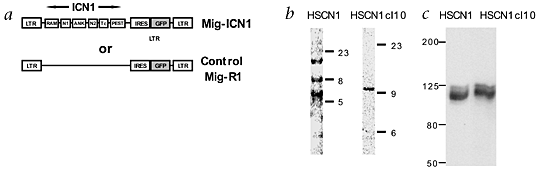
common lymphoid precursor16,17 and promotes expansion of CD8+ cells18,19,20. Here, we demonstrate that constitutive Notch1 signaling in hematopoietic cells established immortalized,
cytokine-dependent cell lines that generated progeny with either lymphoid or myeloid characteristics both _in vitro_ and _in vivo_. These data support a role for Notch signaling in
regulating hematopoietic stem cell self-renewal. Furthermore, the establishment of clonal, pluripotent cell lines provides the opportunity to assess mechanisms regulating stem cell
commitment and demonstrates a general method for immortalizing stem cell populations for further analysis. Access through your institution Buy or subscribe This is a preview of subscription
content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue
Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL
ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS NLRC3 SIGNALING IS INDISPENSABLE FOR
HEMATOPOIETIC STEM CELL EMERGENCE VIA NOTCH SIGNALING IN VERTEBRATES Article Open access 03 January 2024 THE UNIVERSAL STEM CELL Article Open access 28 October 2022 SYSTEMATIC IDENTIFICATION
OF CELL-FATE REGULATORY PROGRAMS USING A SINGLE-CELL ATLAS OF MOUSE DEVELOPMENT Article 11 July 2022 REFERENCES * S. Artavanis-Tsakonas, Rand, M.D. & Lake, R.J. Notch signaling: Cell
fate control and signal integration in development. _Science_ 284, 770– 776 (1999). Article CAS Google Scholar * Simpson, P. Introduction: Notch signalling and choice of cell fates in
development. _ Semin. Cell Dev. Biol._ 9, 581–582 (1998). Article CAS Google Scholar * Coffman, C., Harris, W. & Kintner, C. Xotch, the Xenopus homolog of Drosophila notch. _ Science_
249, 1438–1441 ( 1990). Article CAS Google Scholar * Weinmaster, G., Roberts, V.J. & Lemke, G. A homolog of Drosophila Notch expressed during mammalian development. _Development_
113, 199–205 (1991). CAS PubMed Google Scholar * Del Amo, F.F. _ et al_. Expression pattern of Motch, a mouse homolog of Drosophila Notch, suggests an important role in early
postimplantation mouse development . _Development_ 115, 737– 744 (1992). CAS PubMed Google Scholar * Ellisen, L.W. _ et al_. TAN-1, the human homolog of the Drosophila notch gene, is
broken by chromosomal translocations in T lymphoblastic neoplasms. _ Cell_ 66, 649–661 ( 1991). Article CAS Google Scholar * Weinmaster, G., Roberts, V.J. & Lemke, G. Notch2: a second
mammalian Notch gene. _ Development_ 116, 931–941 (1992). CAS Google Scholar * Milner, L.A., Kopan, R., Martin, D.I. & Bernstein, I.D. A human homologue of the Drosophila
developmental gene, Notch, is expressed in CD34+ hematopoietic precursors. _Blood_ 83, 2057– 2062 (1994). CAS PubMed Google Scholar * Ohishi, K. _et al_. Monocytes express high amounts of
Notch and undergo cytokine specific apoptosis following interaction with the Notch ligand, Delta-1. _ Blood_ 95, 2847–2854 ( 2000). CAS PubMed Google Scholar * Varnum-Finney, B. _ et
al_. The Notch ligand, Jagged-1, influences the development of primitive hematopoietic precursor cells. _Blood_ 91, 4084–4091 (1998). CAS Google Scholar * Li, L. et al. The human homolog
of rat Jagged1 expressed by marrow stroma inhibits differentiation of 32D cells through interaction with Notch1. _ Immunity_ 8, 43–55 ( 1998). Article CAS Google Scholar * Jones, P. _et
al_. Stromal expression of Jagged 1 promotes colony formation by fetal hematopoietic progenitor cells. _Blood_ 92, 1505–1511 (1998). CAS Google Scholar * Carlesso, N., Aster, J.C., Sklar,
J. & Scadden, D.T. Notch1-induced delay of human hematopoietic progenitor cell differentiation is associated with altered cell cycle kinetics. _Blood_ 93, 838–848 (1999). CAS Google
Scholar * Han, W., Ye, Q. & Moore, M. A soluble form of human Delta-like-1 inhibits differentiation of hematopoietic progenitor cells. _Blood_ 95, 1616– 1625 (2000). CAS Google Scholar
* Walker, L. _ et al_. The Notch/Jagged pathway inhibits proliferation of human hematopoietic progenitors _in vitro_. _Stem Cells_ 17, 162–171 (1999). Article CAS Google Scholar *
Radtke, F. _ et al_. Deficient T cell fate specification in mice with an induced inactivation of Notch1. _Immunity_ 10, 547 –558 (1999). Article CAS Google Scholar * Pui, J.C. _et al_.
Notch1 expression in early lymphopoiesis influences B versus T lineage determination. _Immunity_ 11, 299 –308 (1999). Article CAS Google Scholar * Robey, E. _et al_. An activated form of
Notch influences the choice between CD4 and CD8 T cell lineages. _Cell_ 87, 483– 492 (1996). Article CAS Google Scholar * Yasutomo, K., Doyle, C., Miele, L. & Germain, R.N. The
duration of antigen receptor signalling determines CD4+ versus CD8+ T-cell lineage fate. _Nature_ 404, 506–510 (2000). Article CAS Google Scholar * Deftos, M.L., Huang, E., Ojala, E.W.,
Forbush, K.A. & Bevan, M.J. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. _Immunity_ 13, 73–84 (2000). Article CAS Google Scholar * Tsai, S., Bartelmez, S.,
Sitnicka, E. & Collins, S. Lymphohematopoietic progenitors immortalized by a retroviral vector harboring a dominant-negative retinoic acid receptor can recapitulate lymphoid, myeloid,
and erythroid development. _Genes Dev._ 8, 2831–2841 (1994). Article CAS Google Scholar * Nutt, S.L., Heavey, B., Rolink, A.G. & Busslinger, M. Commitment to the B-lymphoid lineage
depends on the transcription factor Pax5 . _Nature_ 401, 556–562 (1999). Article CAS Google Scholar * Rolink, A.G., Nutt, S.L., Melchers, F. & Busslinger, M. Long-term in vivo
reconstitution of T-cell development by Pax5-deficient B-cell progenitors. _Nature_ 401, 603– 606 (1999). Article CAS Google Scholar * Capobianco, A.J., Zagouras, P., Blaumueller, C.M.,
Artavanis-Tsakonas, S. & Bishop, J.M. Neoplastic transformation b truncated alleles of human Notch1/Tan1 and Notch2 . _Mol. Cell Biol._ 17, 6265– 6273 (1997). Article CAS Google
Scholar * Kiem, H.P. _ et al_. Improved gene transfer into baboon marrow repopulating cells using recombinant human fibronectin fragment CH-296 in combination with interleukin-6, stem cell
factor, FLT-3 ligand, and megakaryocyte growth and development factor . _Blood_ 92, 1878–1886 (1998). CAS PubMed Google Scholar * Pear, W.S. _ et al_. Efficient and rapid induction of a
CML-like myeloproliferative disease in mice receiving BCR/abl transduced bone marrow. _Blood _ 92, 3780–3792 ( 1998). CAS Google Scholar * Huppert, S. _ et al_. Embryonic lethality in mice
homozygous for a processing deficient Notch1 allele. _Nature_ 405, 966– 970 (2000). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We thank M. Linenberger, S. Collins
and K. Ohishi for criticism of the manuscript. This work was supported by grants P50 HL54881, RO1-CA82308-01 and RO1-AI47833-01 from the National Institutes of Health. I.D.B is also
supported as an American Cancer Society-F.M. KirbyClinical Research Professorship, and W.S.P is the recipient of a Scholar Award from the Leukemia and Lymphoma Society. AUTHOR INFORMATION
AUTHORS AND AFFILIATIONS * Clinical Research Division, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave. N., D2-373, Seattle, 98109, Washington, USA Barbara Varnum-Finney, Carolyn
Brashem-Stein, Cynthia Nourigat, David Flowers & Irwin D. Bernstein * Department of Pathology and Institute for Medicine and Engineering, 611 BRB II/III, 421 Curie Blvd. Lanwei Xu, Sonia
Bakkour & Warren S. Pear * University of Pennsylvania, Philadelphia, 19104, Pennsylvania, USA Lanwei Xu, Sonia Bakkour & Warren S. Pear * The Department of Pediatrics, The
University of Washington, Seattle, 98105, Washington, USA Irwin D. Bernstein Authors * Barbara Varnum-Finney View author publications You can also search for this author inPubMed Google
Scholar * Lanwei Xu View author publications You can also search for this author inPubMed Google Scholar * Carolyn Brashem-Stein View author publications You can also search for this author
inPubMed Google Scholar * Cynthia Nourigat View author publications You can also search for this author inPubMed Google Scholar * David Flowers View author publications You can also search
for this author inPubMed Google Scholar * Sonia Bakkour View author publications You can also search for this author inPubMed Google Scholar * Warren S. Pear View author publications You can
also search for this author inPubMed Google Scholar * Irwin D. Bernstein View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR
Correspondence to Barbara Varnum-Finney. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Varnum-Finney, B., Xu, L., Brashem-Stein, C. _et al._
Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. _Nat Med_ 6, 1278–1281 (2000). https://doi.org/10.1038/81390 Download citation *
Received: 15 June 2000 * Accepted: 27 September 2000 * Issue Date: November 2000 * DOI: https://doi.org/10.1038/81390 SHARE THIS ARTICLE Anyone you share the following link with will be able
to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative