Play all audios:
Access through your institution Buy or subscribe To the Editor: Scientists commonly form histograms of counted events from their data, and extract parameters by fitting to a known model.
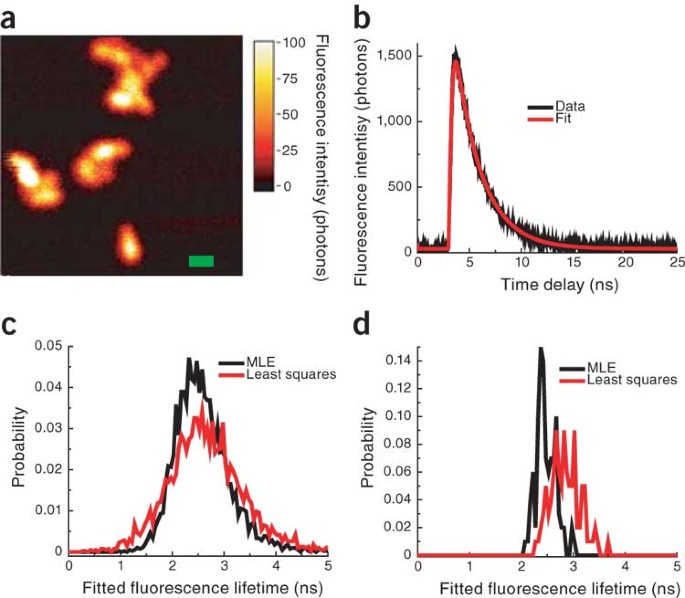
Anytime a scientist counts photons, molecules, cells or data for individuals in histogram bins and fits that to a distribution, he or she is fitting an event-counting histogram. Here we aim
to convince the scientific community to use the maximum likelihood estimator (MLE) for Poisson deviates when fitting event-counting histograms rather than the typically used least-squares
measure. We describe how to use the MLE to fit data efficiently and robustly, and provide example code (Supplementary Software). This is a preview of subscription content, access via your
institution RELEVANT ARTICLES Open Access articles citing this article. * GLOBAL FITTING FOR HIGH-ACCURACY MULTI-CHANNEL SINGLE-MOLECULE LOCALIZATION * Yiming Li * , Wei Shi * … Jonas Ries
_Nature Communications_ Open Access 06 June 2022 * GENERATIVE ADVERSARIAL NETWORK ENABLES RAPID AND ROBUST FLUORESCENCE LIFETIME IMAGE ANALYSIS IN LIVE CELLS * Yuan-I Chen * , Yin-Jui Chang
* … Hsin-Chih Yeh _Communications Biology_ Open Access 11 January 2022 * CORRECTING ARTIFACTS IN SINGLE MOLECULE LOCALIZATION MICROSCOPY ANALYSIS ARISING FROM PIXEL QUANTUM EFFICIENCY
DIFFERENCES IN SCMOS CAMERAS * Hazen P. Babcock * , Fang Huang * & Colenso M. Speer _Scientific Reports_ Open Access 02 December 2019 ACCESS OPTIONS Access through your institution
Subscribe to this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full
article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs *
Contact customer support REFERENCES * Ross, S.M. _Introduction to Probability and Statistics for Engineers and Scientists_ (Wiley, New York, New York, 1987). Google Scholar * Turton, D.A.,
Reid, G.D. & Beddard, G.S. _Anal. Chem._ 75, 4182–4187 (2003). Article CAS Google Scholar * Marquardt, D.W. _J. Soc. Indust. Appl. Math._ 11, 431–441 (1963). Article Google Scholar
* Bunch, D.S., Gay, D.M. & Welsch, R.E. _ACM T. Math. Software_ 19, 109–130 (1993). Article Google Scholar * Gay, D.M. & Welsch, R.E. _J. Am. Stat. Assoc._ 83, 990–998 (1988).
Article Google Scholar * Nishimura, G. & Tamura, M. _Phys. Med. Biol._ 50, 1327–1342 (2005). Article Google Scholar Download references ACKNOWLEDGEMENTS This work was performed under
the auspices of the US Department of Energy by Lawrence Livermore National Laboratory under contract DE-AC52-07NA27344. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Physical and Life
Sciences Directorate, Lawrence Livermore National Laboratory, Livermore, California, USA Ted A Laurence & Brett A Chromy Authors * Ted A Laurence View author publications You can also
search for this author inPubMed Google Scholar * Brett A Chromy View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to
Ted A Laurence. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Note (PDF
870 kb) SUPPLEMENTARY SOFTWARE C code for Levenberg-Marquardt minimization of MLE. (ZIP 17 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Laurence,
T., Chromy, B. Efficient maximum likelihood estimator fitting of histograms. _Nat Methods_ 7, 338–339 (2010). https://doi.org/10.1038/nmeth0510-338 Download citation * Issue Date: May 2010
* DOI: https://doi.org/10.1038/nmeth0510-338 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative