Play all audios:
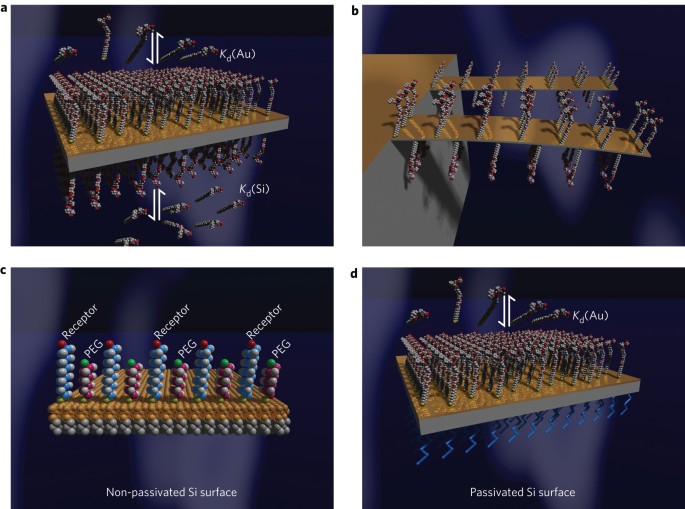
ABSTRACT Cantilever arrays have been used to monitor biochemical interactions and their associated stress. However, it is often necessary to passivate the underside of the cantilever to
prevent unwanted ligand adsorption, and this process requires tedious optimization. Here, we show a way to immobilize membrane receptors on nanomechanical cantilevers so that they can
function without passivating the underlying surface. Using equilibrium theory, we quantitatively describe the mechanical responses of vancomycin, human immunodeficiency virus type 1 antigens
and coagulation factor VIII captured on the cantilever in the presence of competing stresses from the top and bottom cantilever surfaces. We show that the area per receptor molecule on the
cantilever surface influences ligand–receptor binding and plays an important role on stress. Our results offer a new way to sense biomolecules and will aid in the creation of ultrasensitive
biosensors. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe
to this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF
Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact
customer support SIMILAR CONTENT BEING VIEWED BY OTHERS COMPUTATIONAL DESIGN OF SINGLE-STRANDED DNA HAIRPIN APTAMERS IMMOBILIZED ON A BIOSENSOR SUBSTRATE Article Open access 26 May 2021 LOW
COST AND MASSIVELY PARALLEL FORCE SPECTROSCOPY WITH FLUID LOADING ON A CHIP Article Open access 10 November 2022 COOPERATIVE CONTROL OF A DNA ORIGAMI FORCE SENSOR Article Open access 19
February 2024 REFERENCES * Son, K., Guasto, J. S. & Stocker, R. Bacteria can exploit a flagellar buckling instability to change direction. _Nature Phys._ 9, 494–498 (2013). Article CAS
Google Scholar * Lele, P. P., Hosu, B. G. & Berg, H. C. Dynamics of mechanosensing in the bacterial flagellar motor. _Proc. Natl Acad. Sci. USA_ 110, 11839–11844 (2013). Article CAS
Google Scholar * Gebhardt, J. C. & Rief, M. Biochemistry force signaling in biology. _Science_ 324, 1278–1280 (2009). Article CAS Google Scholar * Arlett, J. L., Myers, E. B. &
Roukes, M. L. Comparative advantages of mechanical biosensors. _Nature Nanotech._ 6, 203–215 (2011). Article CAS Google Scholar * Müller, D. J. & Dufrêne, Y. F. Atomic force
microscopy as a multifunctional molecular toolbox in nanobiotechnology. _Nature Nanotech._ 3, 261–269 (2008). Article Google Scholar * Huber, F., Lang, H. P., Backmann, N., Rimoldi, D.
& Gerber, C. Direct detection of a BRAF mutation in total RNA from melanoma cells using cantilever arrays. _Nature Nanotech._ 8, 125–129 (2013). Article CAS Google Scholar * McKendry,
R. A. et al. Multiple label-free biodetection and quantitative DNA-binding assays on a nanomechanical cantilever array. _Proc. Natl Acad. Sci. USA_ 99, 9783–9788 (2002). Article CAS
Google Scholar * Wu, G. H. et al. Bioassay of prostate-specific antigen (PSA) using microcantilevers. _Nature Biotechnol._ 19, 856–860 (2001). Article CAS Google Scholar * Fritz, J. et
al. Translating biomolecular recognition into nanomechanics. _Science_ 288, 316–318 (2000). Article CAS Google Scholar * Berger, R. et al. Surface stress in the self-assembly of
alkanethiols on gold. _Science_ 276, 2021–2024 (1997). Article CAS Google Scholar * Gfeller, K. Y., Nugaeva, N. & Hegner, M. Micromechanical oscillators as rapid biosensor for the
detection of active growth of _Escherichia coli_. _Biosens. Bioelectron._ 21, 528–533 (2005). Article CAS Google Scholar * Longo, G. et al. Rapid detection of bacterial resistance to
antibiotics using AFM cantilevers as nanomechanical sensors. _Nature Nanotech_. 8, 522–526 (2013). Article CAS Google Scholar * Ndieyira, J. W. et al. Surface-stress sensors for rapid and
ultrasensitive detection of active free drugs in human serum. _Nature Nanotech_. 9, 225–232 (2014). Article CAS Google Scholar * Ndieyira, J. W. et al. Nanomechanical detection of
antibiotic–mucopeptide binding in a model for superbug drug resistance. _Nature Nanotech._ 3, 691–696 (2008). Article CAS Google Scholar * Dupres, V. et al. Nanoscale mapping and
functional analysis of individual adhesins on living bacteria. _Nature Methods_ 2, 515–520 (2001). Article Google Scholar * Zhang, J. et al. Rapid and label-free nanomechanical detection
of biomarker transcripts in human RNA. _Nature Nanotech._ 1, 214–220 (2006). Article CAS Google Scholar * Kosaka, P. M. et al. Detection of cancer biomarkers in serum using a hybrid
mechanical and optoplasmonic nanosensor. _Nature Nanotech_. 9, 1047–1053 (2014). Article CAS Google Scholar * Boisen, A., Dohn, S., Keller, S. S., Schmid, S. & Tenje, M.
Cantilever-like micromechanical sensors. _Rep. Prog. Phys._ 74, 036101 (2011). Article Google Scholar * Burg, T. P. et al. Weighing of biomolecules, single cells and single nanoparticles
in fluid. _Nature_ 446, 1066–1069 (2007). Article CAS Google Scholar * Shu, W. et al. DNA molecular motor driven micromechanical cantilever arrays. _J. Am. Chem. Soc_. 127, 17054–17060
(2005). Article CAS Google Scholar * Backmann, N. et al. A label-free immunosensor array using single-chain antibody fragments. _Proc. Natl Acad. Sci. USA_ 102, 14587–14592 (2005).
Article CAS Google Scholar * Mukhopadhyay, R. et al. Cantilever sensor for nanomechanical detection of specific protein conformations. _Nano Lett_. 5, 2385–2388 (2005). Article CAS
Google Scholar * Nieto, M. & Perkins, H. R. Modifications of acyl–D-alanyl–D-alanine terminus affecting complex-formation with vancomycin. _Biochem. J_. 123, 773–787 (1971). Article
CAS Google Scholar * Williams, D. H., Maguire, A. J., Tsuzuki, W. & Westwell, M. S. An analysis of the origins of a cooperative binding energy of dimerisation. _Science_ 280, 711–714
(1998). Article CAS Google Scholar * Sieradzki, K. et al. The development of vancomycin resistance in a patient with methicillin-resistant _Staphylococcus aureus_ infection. _N. Engl. J.
Med._ 340, 517–523 (1999). Article CAS Google Scholar * Barna, J. C. & Williams, D. H. The structure and mode of action of glycopeptides antibiotics of the vancomycin group. _Annu.
Rev. Microbiol_. 38, 339–357 (1984). Article CAS Google Scholar * West, R. Inorganic chemistry: two-armed silicon. _Nature_ 485, 49–50 (2012). Article CAS Google Scholar * Hamers, R.
J. Formation and characterization of organic monolayers on semiconductor surfaces. _Annu. Rev. Anal. Chem_. 1, 707–736 (2008). Article CAS Google Scholar * Haas, A. The chemistry of
silicon–sulfur compounds. _Angew. Chem. Int. Ed. Engl._ 4, 1014–1023 (1965). Article CAS Google Scholar * Love, J. C. et al. Self-assembled monolayers of thiolates on metals as a form of
nanotechnology. _Chem. Rev_. 289, 1103–1169 (2005). Article Google Scholar * Kolomenskii, A. A., Gershon, P. D. & Schuessler, H. A. Sensitivity and detection limit of concentration and
adsorption measurements by laser-induced surface-plasmon resonance. _Appl. Opt._ 36, 6539–6547 (1997). Article CAS Google Scholar * Stenberg, E., Persson, B., Roos, H. & Urbaniczky,
C. Quantitative determination of surface concentration of protein surface plasmon resonance using radiolabeled proteins. _J. Colloid Interface Sci._ 143, 513–526 (1991). Article CAS Google
Scholar * Morita, M., Ohmi, T., Hasegawa, E., Kawakami, M. & Ohwada, M. Growth of native oxide on a silicon surface. _J. Appl. Phys._ 68, 1272–1281 (1990). Article CAS Google Scholar
* Prime, K. L. & Whitesides, G. M. Self-assembled organic monolayers—model systems for studying adsorption of proteins at surfaces. _Science_ 252, 1164–1167 (1991). Article CAS
Google Scholar * Prime, K. L. & Whitesides, G. M. Adsorption of proteins onto surfaces containing end-attached oligo(ethylene oxide)—a model system using self-assembled monolayers. _J.
Am. Chem. Soc._ 115, 10714–10721 (1993). Article CAS Google Scholar * Hamers-Casterman, C. et al. Naturally occurring antibodies devoid of light chains. _Nature_ 363, 446–448 (1993).
Article CAS Google Scholar * Hultberg, A. et al. Lactobacillli expressing llama VHH fragments neutralise _Lactococcus_ phages. _BMC Biotechnology_ 84, 58 (2007). Article Google Scholar
* Strokappe, N. et al. Llama antibodies recognizing various epitopes of the CD4bs neutralize a broad range of HIV-1 subtypes A, B, and C. _PLoS One_ 7, e33298 (2012). Article CAS Google
Scholar * McCoy, L. E. et al. Potent and broad neutralization of HIV-1 by a llama antibody elicited by immunization. _J. Exp. Med._ 209, 1091–1103 (2012). Article CAS Google Scholar *
McCoy, L. E. et al. Molecular evolution of broadly neutralizing llama antibodies to the CD4-binding site of HIV-1. _PLoS Pathog_ 10, e1004552 (2014). Article Google Scholar * McCoy, L. E.
et al. Broadly neutralizing VHH against HIV-1. Patent application WO 2013036130 A1 (EP2753644A1, US201501158934). * Zhou, F. et al. Sensitive sandwich ELISA based on a gold nanoparticle
layer for cancer detection. _Analyst_ 137, 1779–1784 (2012). Article CAS Google Scholar * Liu, J., Bartesaghi, A., Borgnia, M. J., Sapiro, G. & Subramaniam, S. Molecular architecture
of native HIV-1 gp120 trimers. _Nature_ 455, 109–113 (2008). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We thank the EPSRC Grand Challenge in Nanotechnology for
Healthcare (EP/G0620064/1), I-sense EPSRC IRC in Early Warning Sensing Systems for Infectious Diseases (EP/G062064/1), Royal Society (RS), Targanta Therapeutics, Bio Nano Consulting (BNC),
the European Union FP7 Project VSMMART Nano (managed by BNC) for funding. The authors also thank J. Russat (London Centre for Nanotechnology), S. Sivachelvam (London Centre for
Nanotechnology), M. Rehak (Sphere Fluidics, UK), R.A. Weiss (University College London) C.T. Verrips (QVQuality, Utrecht), T. Philips (Utrecht University), M. Morfini (University of
Florence), T. Cass (Imperial College), V. Emery (Surrey Business School) and G. Aeppli (Paul Scherrer Institut) for the kind gift of materials and for helpful discussions. The glycoprotein
antigens (gp140CN54 and gp140UG37) to llama antibody fragments were provided by the Centre for AIDS Reagents, National Institute for Biological Standards and Control (NIBSC) of the UK
Medicines & Healthcare Products Regulatory Agency (MHRA). AUTHOR INFORMATION Author notes * Samadhan B. Patil, Manuel Vögtli, Benjamin Webb and Joseph W. Ndieyira: These authors
contributed equally to this work AUTHORS AND AFFILIATIONS * London Centre for Nanotechnology and Departments of Medicine and Physics, University College London, 17–19 Gordon Street, London,
WC1H 0AH, UK Samadhan B. Patil, Manuel Vögtli, Benjamin Webb, Rachel A. McKendry & Joseph W. Ndieyira * Department of Materials, Imperial College London, London, SW7 2AZ, UK Samadhan B.
Patil & Yeong-Ah Soh * Division of Infection & Immunity, University College London, Cruciform Building, Gower Street, London, WC1E 6BT, UK Benjamin Webb * UCL Institute for Liver and
Digestive Health, Royal Free Hospital, London, NW3 2QG, UK Giuseppe Mazza & Massimo Pinzani * Department of Chemistry, Jomo Kenyatta University of Agriculture and Technology, Nairobi,
PO Box 62000, Kenya Joseph W. Ndieyira Authors * Samadhan B. Patil View author publications You can also search for this author inPubMed Google Scholar * Manuel Vögtli View author
publications You can also search for this author inPubMed Google Scholar * Benjamin Webb View author publications You can also search for this author inPubMed Google Scholar * Giuseppe Mazza
View author publications You can also search for this author inPubMed Google Scholar * Massimo Pinzani View author publications You can also search for this author inPubMed Google Scholar *
Yeong-Ah Soh View author publications You can also search for this author inPubMed Google Scholar * Rachel A. McKendry View author publications You can also search for this author inPubMed
Google Scholar * Joseph W. Ndieyira View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS J.W.N. designed the experiments. J.W.N. and M.V.
performed the experiments on antibiotics. J.W.N., S.B.P. and B.W. performed the experiments on HIV antigen detections. J.W.N. and S.B.P. formulated the mathematical model to decouple
competing surface binding kinetics at Au (top surface of cantilever) and Si (bottom surface of cantilever). J.W.N. performed the experiments on blood clotting proteins and wrote the paper.
All authors discussed the results and commented on the manuscript. CORRESPONDING AUTHOR Correspondence to Joseph W. Ndieyira. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no
competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary information (PDF 452 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE
CITE THIS ARTICLE Patil, S., Vögtli, M., Webb, B. _et al._ Decoupling competing surface binding kinetics and reconfiguration of receptor footprint for ultrasensitive stress assays. _Nature
Nanotech_ 10, 899–907 (2015). https://doi.org/10.1038/nnano.2015.174 Download citation * Received: 15 November 2014 * Accepted: 06 July 2015 * Published: 17 August 2015 * Issue Date: October
2015 * DOI: https://doi.org/10.1038/nnano.2015.174 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link
is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative