Play all audios:
We recently reported that damaging _de novo_ mutations in persons with schizophrenia from otherwise healthy families disrupt genes that orchestrate neurogenesis in fetal prefrontal cortex
(Gulsuner et al, 2013). By sequencing genomic DNA from entire families, we identified point mutations and copy number variants that appeared _de novo_ in 105 persons with schizophrenia and
in 84 of their healthy siblings. Patients were more likely than unaffected siblings to harbor damaging _de novo_ mutations (47/105 _vs_ 30/84, OR=1.91, X2=4.45; _p_=0.035). The proportion of
schizophrenia attributable to damaging _de novo_ mutations in this sample was 21%. Even more striking than the difference in mutation frequencies was the distinctive functional relationship
among the genes harboring mutations. The genes disrupted by damaging _de novo_ mutations in patients formed a network defined by protein interaction (Mostafavi et al, 2008) and by
transcriptional co-expression (BrainSpan: Atlas of the Developing Human Brain, 2013) in the dorsolateral and ventrolateral prefrontal cortex during fetal development. Interactions among
these genes were not significantly enriched in other brain regions or during other developmental periods, nor were interactions enriched among genes harboring benign _de novo_ events. Of the
54 genes disrupted by damaging _de novo_ mutations in patients, 50 mapped to this network. These genes are active in pathways critical to neurogenesis, including neuronal migration,
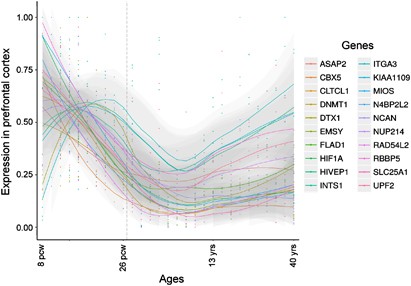
synaptic transmission, signaling, transcriptional regulation, and transport. Many network genes share a pattern of high expression in prefrontal cortex during the early fetal development,
with transcription levels declining in the late fetal development and childhood only to rise again during young adulthood (Figure 1), coinciding with the typical age of onset of the illness.
The results suggest that aberrant prefrontal cortical development is critical to the pathogenesis of schizophrenia. The prefrontal cortex is highly interconnected with other brain regions,
and organizes information to coordinate executive skills, including working memory, attention span, problem solving, and self-regulation (Catts et al, 2013). Given the clearinghouse role of
prefrontal cortex in brain function, disruptions in this region during early development likely result in the overall loss of neocortical integrity and connectivity. Impairments in executive
functions are characteristic of schizophrenia, suggesting that higher-order cognitive deficits are early manifestations of the syndrome, with psychosis emerging as critical brain regions
mature. Some genes with _de novo_ mutations in patients function in neurotransmitter pathways that suggest possible avenues for treatment, including GLS (glutamate synthesis), ADCY9
(glutamate and GABA signaling), and SLC18A2 (serotonin, dopamine, norepinephrine, epinephrine, and histamine transport). _CACNA1I_, a brain-specific, T-type calcium channel that regulates
neuronal firing in the thalamus, striatum, nucleus accumbens, and prefrontal cortex, was the only gene disrupted in two unrelated probands, each harboring a different _de novo_ event. Some
antipsychotic medications, including clozapine, inhibit T-type calcium channels (Choi and Rhim, 2010). Key mechanisms underlying complex psychiatric disorders can be identified by
characterizing brain pathways disrupted by mutant genes in affected persons. By integrating genomic analyses with brain mapping strategies, we were able to define possible disease-related
processes and to identify potential targets for treatment. Applying the same approach in large-scale sequencing studies will help elucidate basic neurobiological mechanisms underlying normal
brain circuitry and neuropsychiatric disease. FUNDING AND DISCLOSURE The authors declare no conflict of interest. REFERENCES * BrainSpan: Atlas of the Developing Human Brain (2013)).
www.brainspan.org. * Catts VS, Fung SJ, Long LE, Joshi D, Vercammen A, Allen KM _et al_ (2013). Rethinking schizophrenia in the context of normal neurodevelopment. _Front Cell Neurosci_ 7:
60. Article Google Scholar * Choi KH, Rhim H (2010). Inhibition of recombinant Ca(v)3.1 (alpha1G) T-type calcium channels by the antipsychotic drug clozapine. _Eur J Pharmacol_ 626:
123–130. Article CAS Google Scholar * Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S _et al_ (2013). Spatial and temporal mapping of _de novo_ mutations in schizophrenia to
a fetal prefrontal cortical network. _Cell_ 154: 518–529. Article CAS Google Scholar * Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q (2008). GeneMANIA: a real-time multiple
association network integration algorithm for predicting gene function. _Genome Biol_ 9 (Suppl 1): S4. Article Google Scholar Download references ACKNOWLEDGEMENTS This work was supported
by NIMH grants R01MH083989, R01MH083849, R01MH083756, R01MH084071, and R03TW008696. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Medicine, University of Washington, Seattle,
WA, USA Suleyman Gulsuner * Department of Genome Sciences, University of Washington, Seattle, WA, USA Suleyman Gulsuner * Department of Psychiatry, University of Washington, Seattle, WA, USA
Jon M McClellan Authors * Suleyman Gulsuner View author publications You can also search for this author inPubMed Google Scholar * Jon M McClellan View author publications You can also
search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Jon M McClellan. POWERPOINT SLIDES POWERPOINT SLIDE FOR FIG. 1 RIGHTS AND PERMISSIONS Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Gulsuner, S., McClellan, J. _De Novo_ Mutations in Schizophrenia Disrupt Genes Co-Expressed in Fetal Prefrontal Cortex.
_Neuropsychopharmacol_ 39, 238–239 (2014). https://doi.org/10.1038/npp.2013.219 Download citation * Published: 09 December 2013 * Issue Date: January 2014 * DOI:
https://doi.org/10.1038/npp.2013.219 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative