Play all audios:
ABSTRACT During the development of an organism, cells are exposed to a myriad of signals, structural components and scaffolds, which collectively make up the cellular microenvironment. The
majority of current developmental biology studies examine the effect of individual or small subsets of molecules and parameters on cellular behavior, and they consequently fail to explore
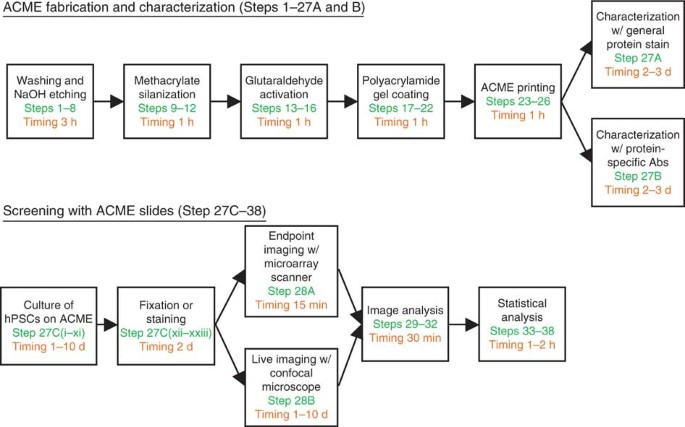
the complexity of factors to which cells are exposed. Here we describe a technology, referred to as arrayed cellular microenvironments (ACMEs), that allows for a high-throughput examination
of the effects of multiple extracellular components in a combinatorial manner on any cell type of interest. We will specifically focus on the application of this technology to human
pluripotent stem cells (hPSCs), a population of cells with tremendous therapeutic potential, and one for which growth and differentiation conditions are poorly characterized and far from
defined and optimized. A standard ACME screen uses the technologies previously applied to the manufacture and analysis of DNA microarrays, requires standard cell-culture facilities and can
be performed from beginning to end within 5–10 days. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS
Access through your institution Subscribe to this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on
SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about
institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS GENOME-WIDE POOLED CRISPR SCREENING IN NEUROSPHERES Article 07 June 2023
CELLULAR POPULATION DYNAMICS SHAPE THE ROUTE TO HUMAN PLURIPOTENCY Article Open access 17 May 2023 A 3D SYSTEM TO MODEL HUMAN PANCREAS DEVELOPMENT AND ITS REFERENCE SINGLE-CELL TRANSCRIPTOME
ATLAS IDENTIFY SIGNALING PATHWAYS REQUIRED FOR PROGENITOR EXPANSION Article Open access 25 May 2021 REFERENCES * Chen, Y., Yu, P., Luo, J. & Jiang, Y. Secreted protein prediction system
combining CJ-SPHMM, TMHMM, and PSORT. _Mamm. Genome_ 14, 859–865 (2003). Article CAS Google Scholar * Grimmond, S.M. et al. The mouse secretome: functional classification of the proteins
secreted into the extracellular environment. _Genome Res._ 13, 1350–1359 (2003). Article CAS Google Scholar * Brafman, D.A., Shah, K.D., Fellner, T., Chien, S. & Willert, K. Defining
long-term maintenance conditions of human embryonic stem cells with arrayed cellular microenvironment technology. _Stem Cells Dev._ 18, 1141–1154 (2009). Article Google Scholar * Brafman,
D.A. et al. Investigating the role of the extracellular environment in modulating hepatic stellate cell biology with arrayed combinatorial microenvironments. _Integr. Biol. (Camb)_ 1,
513–524 (2009). Article CAS Google Scholar * Flaim, C.J., Chien, S. & Bhatia, S.N. An extracellular matrix microarray for probing cellular differentiation. _Nat. Methods_ 2, 119–125
(2005). Article CAS Google Scholar * Flaim, C.J., Teng, D., Chien, S. & Bhatia, S.N. Combinatorial signaling microenvironments for studying stem cell fate. _Stem Cells Dev._ 17, 29–39
(2008). Article CAS Google Scholar * Soen, Y., Mori, A., Palmer, T.D. & Brown, P.O. Exploring the regulation of human neural precursor cell differentiation using arrays of signaling
microenvironments. _Mol. Syst. Biol._ 2, 37 (2006). Article Google Scholar * La Barge, M.A. et al. Human mammary progenitor cell fate decisions are products of interactions with
combinatorial microenvironments. _Integr. Biol. (Camb)_ 1, 70–79 (2009). Article CAS Google Scholar * Brafman, D.A. et al. Long-term human pluripotent stem cell self-renewal on synthetic
polymer surfaces. _Biomaterials_ 31, 9135–9144 (2010). Article CAS Google Scholar * Anderson, D.G., Putnam, D., Lavik, E.B., Mahmood, T.A. & Langer, R. Biomaterial microarrays: rapid,
microscale screening of polymer-cell interaction. _Biomaterials_ 26, 4892–4897 (2005). Article CAS Google Scholar * Damoiseaux, R., Sherman, S.P., Alva, J.A., Peterson, C. & Pyle,
A.D. Integrated chemical genomics reveals modifiers of survival in human embryonic stem cells. _Stem Cells_ 27, 533–542 (2009). Article CAS Google Scholar * Watanabe, K. et al. A ROCK
inhibitor permits survival of dissociated human embryonic stem cells. _Nat. Biotechnol._ 25, 681–686 (2007). Article CAS Google Scholar * Jones, C.N. et al. Multifunctional protein
microarrays for cultivation of cells and immunodetection of secreted cellular products. _Anal. Chem._ 80, 6351–6357 (2008). Article CAS Google Scholar * Fernandes, T.G. et al. On-chip,
cell-based microarray immunofluorescence assay for high-throughput analysis of target proteins. _Anal. Chem._ 80, 6633–6639 (2008). Article CAS Google Scholar * Hariharan, R. The analysis
of microarray data. _Pharmacogenomics_ 4, 477–497 (2003). Article CAS Google Scholar * Svrakic, N.M., Nesic, O., Dasu, M.R., Herndon, D. & Perez-Polo, J.R. Statistical approach to
DNA chip analysis. _Recent Prog. Horm. Res._ 58, 75–93 (2003). Article CAS Google Scholar * Shannon, W., Culverhouse, R. & Duncan, J. Analyzing microarray data using cluster analysis.
_Pharmacogenomics_ 4, 41–52 (2003). Article CAS Google Scholar * Malo, N., Hanley, J.A., Cerquozzi, S., Pelletier, J. & Nadon, R. Statistical practice in high-throughput screening
data analysis. _Nat. Biotechnol._ 24, 167–175 (2006). Article CAS Google Scholar * Makarenkov, V. et al. HTS-Corrector: software for the statistical analysis and correction of
experimental high-throughput screening data. _Bioinformatics_ 22, 1408–1409 (2006). Article CAS Google Scholar * Box, G., Hunter, W.G., Hunter, J.S. & Hunter, W. _Statistics for
Experimenters_ (Wiley, 1978). * Eisen, M.B., Spellman, P.T., Brown, P.O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. _Proc. Natl Acad. Sci. USA_ 95,
14863–14868 (1998). Article CAS Google Scholar * Xu, C. et al. Feeder-free growth of undifferentiated human embryonic stem cells. _Nat. Biotechnol._ 19, 971–974 (2001). Article CAS
Google Scholar * Nandivada, H. et al. Fabrication of synthetic polymer coatings and their use in feeder-free culture of human embryonic stem cells. _Nat. Protoc._ 6, 1037–1043 (2011).
Article CAS Google Scholar * Ware, C.B., Nelson, A.M. & Blau, C.A. A comparison of NIH-approved human ESC lines. _Stem Cells_ 24, 2677–2684 (2006). Article CAS Google Scholar *
Willert, K.H. Isolation and application of bioactive Wnt proteins. _Methods Mol. Biol._ 468, 17–29 (2008). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS D.A.B. was
supported by funding from the University of California San Diego Stem Cell Program and by a gift from Michael and Nancy Kaehr. This research was supported in part by the California Institute
of Regenerative Medicine (RS1-00172-1) to S.C. and (RB1-01406) to K.W. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Cellular and Molecular Medicine, Stem Cell Program, University of
California, San Diego, California, USA David A Brafman & Karl Willert * Department of Bioengineering, University of California, San Diego, California, USA Shu Chien Authors * David A
Brafman View author publications You can also search for this author inPubMed Google Scholar * Shu Chien View author publications You can also search for this author inPubMed Google Scholar
* Karl Willert View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS D.A.B., S.C. and K.W. developed the protocol. D.A.B. and K.W. designed and
performed the experiments. D.A.B., S.C. and K.W. analyzed the results. D.A.B., S.C. and K.W. wrote the manuscript. CORRESPONDING AUTHORS Correspondence to David A Brafman or Karl Willert.
ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIG 1 Schematic representation of ACME format. (a)
Schematic of a 10x10 subarray that contains 20 different conditions spotted in replicates of five. Spot diameter (closed circles) is 150 μm and the center to center distance between
neighboring spots is 450 μm. These subarrays can be arranged to generate several different designs including (b) a 8x2, 1600 spot/320 condition, (c) a 16x4, 6400 spot/1280 condition, or (d)
a 16x5, 8000 spot/1600 condition array. (PPT 415 kb) SUPPLEMENTARY TABLE 1 Example of analysis of raw data generated from the array in Figure 5e. (DOC 1214 kb) RIGHTS AND PERMISSIONS
Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Brafman, D., Chien, S. & Willert, K. Arrayed cellular microenvironments for identifying culture and differentiation
conditions for stem, primary and rare cell populations. _Nat Protoc_ 7, 703–717 (2012). https://doi.org/10.1038/nprot.2012.017 Download citation * Published: 15 March 2012 * Issue Date:
April 2012 * DOI: https://doi.org/10.1038/nprot.2012.017 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable
link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative