Play all audios:
ABSTRACT Targeted nucleases are powerful tools for mediating genome alteration with high precision. The RNA-guided Cas9 nuclease from the microbial clustered regularly interspaced short
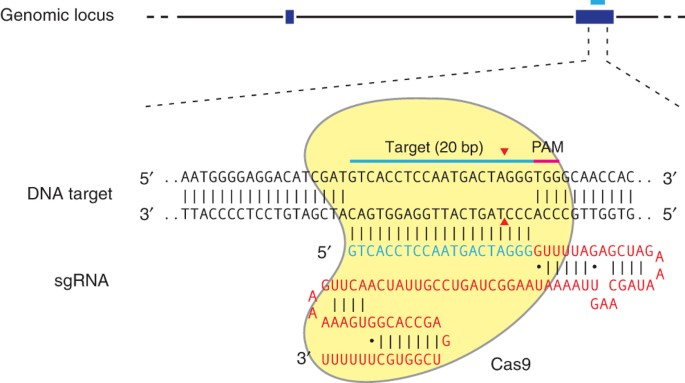
palindromic repeats (CRISPR) adaptive immune system can be used to facilitate efficient genome engineering in eukaryotic cells by simply specifying a 20-nt targeting sequence within its
guide RNA. Here we describe a set of tools for Cas9-mediated genome editing via nonhomologous end joining (NHEJ) or homology-directed repair (HDR) in mammalian cells, as well as generation
of modified cell lines for downstream functional studies. To minimize off-target cleavage, we further describe a double-nicking strategy using the Cas9 nickase mutant with paired guide RNAs.
This protocol provides experimentally derived guidelines for the selection of target sites, evaluation of cleavage efficiency and analysis of off-target activity. Beginning with target
design, gene modifications can be achieved within as little as 1–2 weeks, and modified clonal cell lines can be derived within 2–3 weeks. Access through your institution Buy or subscribe
This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access
$259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are
calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS
CRISPR-CAS12A NUCLEASES FUNCTION WITH STRUCTURALLY ENGINEERED CRRNAS: SYNTHETIC TRACRRNA Article Open access 16 July 2022 EFFICIENT ENGINEERING OF HUMAN AND MOUSE PRIMARY CELLS USING
PEPTIDE-ASSISTED GENOME EDITING Article 24 April 2023 HARNESSING NONCANONICAL CRRNA FOR HIGHLY EFFICIENT GENOME EDITING Article Open access 07 May 2024 REFERENCES * Ding, Q. et al. A TALEN
genome-editing system for generating human stem cell-based disease models. _Cell Stem Cell_ 12, 238–251 (2013). Article CAS Google Scholar * Soldner, F. et al. Generation of isogenic
pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. _Cell_ 146, 318–331 (2011). Article CAS Google Scholar * Carlson, D.F. et al. Efficient
TALEN-mediated gene knockout in livestock. _Proc. Natl. Acad. Sci. USA_ 109, 17382–17387 (2012). Article CAS Google Scholar * Geurts, A.M. et al. Knockout rats via embryo microinjection
of zinc-finger nucleases. _Science_ 325, 433–433 (2009). Article CAS Google Scholar * Takasu, Y. et al. Targeted mutagenesis in the silkworm _Bombyx mori_ using zinc finger nuclease mRNA
injection. _Insect Biochem. Molec._ 40, 759–765 (2010). Article CAS Google Scholar * Watanabe, T. et al. Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger
and TAL effector nucleases. _Nat. Commun._ 3, 1017 (2012). Article Google Scholar * Porteus, M.H. & Baltimore, D. Chimeric nucleases stimulate gene targeting in human cells. _Science_
300, 763 (2003). Article Google Scholar * Miller, J.C. et al. An improved zinc-finger nuclease architecture for highly specific genome editing. _Nat. Biotechnol._ 25, 778–785 (2007).
Article CAS Google Scholar * Sander, J.D. et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). _Nat. Methods_ 8, 67–69 (2011). Article CAS Google
Scholar * Wood, A.J. et al. Targeted genome editing across species using ZFNs and TALENs. _Science_ 333, 307 (2011). Article CAS Google Scholar * Christian, M. et al. Targeting DNA
double-strand breaks with TAL effector nucleases. _Genetics_ 186, 757–761 (2010). Article CAS Google Scholar * Zhang, F. et al. Efficient construction of sequence-specific TAL effectors
for modulating mammalian transcription. _Nat. Biotechnol._ 29, 149–153 (2011). Article Google Scholar * Hockemeyer, D. et al. Genetic engineering of human pluripotent cells using TALE
nucleases. _Nat. Biotechnol._ 29, 731–734 (2011). Article CAS Google Scholar * Reyon, D. et al. FLASH assembly of TALENs for high-throughput genome editing. _Nat. Biotechnol._ 30, 460–465
(2012). Article CAS Google Scholar * Boch, J. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. _Science_ 326, 1509–1512 (2009). Article CAS Google Scholar
* Moscou, M.J. & Bogdanove, A.J. A simple cipher governs DNA recognition by TAL effectors. _Science_ 326, 1501 (2009). Article CAS Google Scholar * Sanjana, N.E. et al. A
transcription activator-like effector toolbox for genome engineering. _Nat. Protoc._ 7, 171–192 (2012). Article CAS Google Scholar * Deveau, H., Garneau, J.E. & Moineau, S. CRISPR/Cas
system and its role in phage-bacteria interactions. _Annu. Rev. Microbiol._ 64, 475–493 (2010). Article CAS Google Scholar * Horvath, P. & Barrangou, R. CRISPR/Cas, the immune system
of bacteria and archaea. _Science_ 327, 167–170 (2010). Article CAS Google Scholar * Makarova, K.S. et al. Evolution and classification of the CRISPR-Cas systems. _Nat. Rev. Microbiol._
9, 467–477 (2011). Article CAS Google Scholar * Bhaya, D., Davison, M. & Barrangou, R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and
regulation. _Annu. Rev. Genet._ 45, 273–297 (2011). Article CAS Google Scholar * Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. _Science_ 339, 819–823 (2013).
Article CAS Google Scholar * Mali, P. et al. RNA-guided human genome engineering via Cas9. _Science_ 339, 823–826 (2013). Article CAS Google Scholar * Jinek, M. et al. RNA-programmed
genome editing in human cells. _eLife_ 2, e00471 (2013). Article Google Scholar * Cho, S.W., Kim, S., Kim, J.M. & Kim, J.S. Targeted genome engineering in human cells with the Cas9
RNA-guided endonuclease. _Nat. Biotechnol._ 31, 230–232 (2013). Article CAS Google Scholar * Garneau, J.E. et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid
DNA. _Nature_ 468, 67–71 (2010). Article CAS Google Scholar * Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. _Science_ 337, 816–821
(2012). Article CAS Google Scholar * Gasiunas, G., Barrangou, R., Horvath, P. & Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity
in bacteria. _Proc. Natl. Acad. Sci. USA_ 109, E2579–E2586 (2012). Article CAS Google Scholar * Urnov, F.D., Rebar, E.J., Holmes, M.C., Zhang, H.S. & Gregory, P.D. Genome editing with
engineered zinc-finger nucleases. _Nat. Rev. Genet._ 11, 636–646 (2010). Article CAS Google Scholar * Hsu, P.D. & Zhang, F. Dissecting neural function using targeted genome
engineering technologies. _ACS Chem. Neurosci._ 3, 603–610 (2012). Article CAS Google Scholar * Perez, E.E. et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing
using zinc-finger nucleases. _Nat. Biotechnol._ 26, 808–816 (2008). Article CAS Google Scholar * Chen, F. et al. High-frequency genome editing using ssDNA oligonucleotides with
zinc-finger nucleases. _Nat. Methods_ 8, 753–755 (2011). Article CAS Google Scholar * Saleh-Gohari, N. & Helleday, T. Conservative homologous recombination preferentially repairs DNA
double-strand breaks in the S phase of the cell cycle in human cells. _Nucleic Acids Res._ 32, 3683–3688 (2004). Article CAS Google Scholar * Marraffini, L.A. & Sontheimer, E.J.
CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. _Science_ 322, 1843–1845 (2008). Article CAS Google Scholar * Brouns, S.J. et al. Small CRISPR RNAs
guide antiviral defense in prokaryotes. _Science_ 321, 960–964 (2008). Article CAS Google Scholar * Barrangou, R. et al. CRISPR provides acquired resistance against viruses in
prokaryotes. _Science_ 315, 1709–1712 (2007). Article CAS Google Scholar * Sapranauskas, R. et al. The _Streptococcus thermophilus_ CRISPR/Cas system provides immunity in _Escherichia
coli_. _Nucleic Acids Res._ 39, 9275–9282 (2011). Article CAS Google Scholar * Magadan, A.H., Dupuis, M.E., Villion, M. & Moineau, S. Cleavage of phage DNA by the _Streptococcus
thermophilus_ CRISPR3-Cas system. _PLoS ONE_ 7, e40913 (2012). Article CAS Google Scholar * Zhang, Y. et al. Processing-Independent CRISPR RNAs limit natural transformation in _Neisseria
meningitidis_. _Mol. Cell_ 50, 488–503 (2013). Article CAS Google Scholar * Hwang, W.Y. et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. _Nat. Biotechnol._ 31,
227–229 (2013). Article CAS Google Scholar * Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. _Cell_ 153,
910–918 (2013). Article CAS Google Scholar * Shen, B. et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. _Cell Res._ 23, 720–723 (2013). Article CAS Google
Scholar * Ran, F.A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. _Cell_ 154, 1380–1389 (2013). Article CAS Google Scholar * Qi, L.S. et al.
Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. _Cell_ 152, 1173–1183 (2013). Article CAS Google Scholar * Chang, N. et al. Genome editing
with RNA-guided Cas9 nuclease in zebrafish embryos. _Cell Res._ 23, 465–472 (2013). Article CAS Google Scholar * Gratz, S.J. et al. Genome engineering of _Drosophila_ with the CRISPR
RNA-guided Cas9 nuclease. _Genetics_ 4, 1029–1035 (2013). Article Google Scholar * Friedland, A.E. et al. Heritable genome editing in _C. elegans_ via a CRISPR-Cas9 system. _Nat. Methods_
10, 741–743 (2013). Article CAS Google Scholar * Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. _Nucleic Acids
Res._ 39, e82 (2011). Article CAS Google Scholar * Schmid-Burgk, J.L., Schmidt, T., Kaiser, V., Honing, K. & Hornung, V. A ligation-independent cloning technique for high-throughput
assembly of transcription activator-like effector genes. _Nat. Biotechnol._ 31, 76–81 (2013). Article CAS Google Scholar * Miller, J.C. et al. A TALE nuclease architecture for efficient
genome editing. _Nat. Biotechnol._ 29, 143–148 (2011). Article CAS Google Scholar * Hsu, P.D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. _Nat. Biotechnol._ 31, 827–832
(2013). Article CAS Google Scholar * Fu, Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. _Nature Biotechnol._ 31, 822–826 (2013). Article
CAS Google Scholar * Tuschl, T. Expanding small RNA interference. _Nat. Biotechnol._ 20, 446–448 (2002). Article CAS Google Scholar * Smithies, O., Gregg, R.G., Boggs, S.S.,
Koralewski, M.A. & Kucherlapati, R.S. Insertion of DNA sequences into the human chromosomal -globin locus by homologous recombination. _Nature_ 317, 230–234 (1985). Article CAS Google
Scholar * Thomas, K.R., Folger, K.R. & Capecchi, M.R. High frequency targeting of genes to specific sites in the mammalian genome. _Cell_ 44, 419–428 (1986). Article CAS Google
Scholar * Hasty, P., Rivera-Perez, J. & Bradley, A. The length of homology required for gene targeting in embryonic stem cells. _Mol. Cell Biol._ 11, 5586–5591 (1991). Article CAS
Google Scholar * Wu, S., Ying, G.X., Wu, Q. & Capecchi, M.R. A protocol for constructing gene targeting vectors: generating knockout mice for the cadherin family and beyond. _Nat.
Protoc._ 3, 1056–1076 (2008). Article CAS Google Scholar * Elliott, B., Richardson, C., Winderbaum, J., Nickoloff, J.A. & Jasin, M. Gene conversion tracts from double-strand break
repair in mammalian cells. _Mol. Cellular Biol._ 18, 93–101 (1998). Article CAS Google Scholar * Guschin, D.Y. et al. A rapid and general assay for monitoring endogenous gene
modification. _Methods Mol. Biol._ 649, 247–256 (2010). Article CAS Google Scholar * Loman, N.J. et al. Performance comparison of benchtop high-throughput sequencing platforms. _Nat.
Biotechnol._ 30, 434–439 (2012). Article CAS Google Scholar * Jiang, W., Bikard, D., Cox, D., Zhang, F. & Marraffini, L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas
systems. _Nat. Biotechnol._ 31, 233–239 (2013). Article CAS Google Scholar * Oliveira, T.Y. et al. Translocation capture sequencing: a method for high throughput mapping of chromosomal
rearrangements. _J. Immunol. Methods_ 375, 176–181 (2012). Article CAS Google Scholar * Gray, S.J. et al. Optimizing promoters for recombinant adeno-associated virus-mediated gene
expression in the peripheral and central nervous system using self-complementary vectors. _Human Gene Ther._ 22, 1143–1153 (2011). Article CAS Google Scholar Download references
ACKNOWLEDGEMENTS We thank B. Holmes for help with computational tools. P.D.H. is a James Mills Pierce Fellow and D.A.S. is a National Science Foundation (NSF) pre-doctoral fellow. V.A. is
supported by NIH Training Grants T32GM007753 and T32GM008313. This work was supported by an NIH Director's Pioneer Award (1DP1-MH100706); an NIH Transformative R01 grant
(1R01-DK097768); the Keck, McKnight, Damon Runyon, Searle Scholars, Vallee, Merkin, Klingenstein and Simons Foundations; Bob Metcalfe; and Jane Pauley. Reagents are available to the academic
community through Addgene and associated protocols; support forums and computational tools are available via the Zhang lab website (http://www.genome-engineering.org/). AUTHOR INFORMATION
Author notes * F Ann Ran and Patrick D Hsu: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * Broad Institute of Massachusetts Institute of Technology (MIT) and
Harvard, Cambridge, Massachusetts, USA F Ann Ran, Patrick D Hsu, Jason Wright, Vineeta Agarwala, David A Scott & Feng Zhang * McGovern Institute for Brain Research, Cambridge,
Massachusetts, USA F Ann Ran, Patrick D Hsu, David A Scott & Feng Zhang * Department of Brain and Cognitive Sciences, MIT, Cambridge, Massachusetts, USA., F Ann Ran, Patrick D Hsu, David
A Scott & Feng Zhang * Department of Biological Engineering, MIT, Cambridge, Massachusetts, USA F Ann Ran, Patrick D Hsu, David A Scott & Feng Zhang * Department of Molecular and
Cellular Biology, Harvard University, Cambridge, Massachusetts, USA F Ann Ran & Patrick D Hsu * Program in Biophysics, Harvard University, MIT, Cambridge, Massachusetts, USA Vineeta
Agarwala * Harvard-MIT Division of Health Sciences and Technology, MIT, Cambridge, Massachusetts, USA Vineeta Agarwala Authors * F Ann Ran View author publications You can also search for
this author inPubMed Google Scholar * Patrick D Hsu View author publications You can also search for this author inPubMed Google Scholar * Jason Wright View author publications You can also
search for this author inPubMed Google Scholar * Vineeta Agarwala View author publications You can also search for this author inPubMed Google Scholar * David A Scott View author
publications You can also search for this author inPubMed Google Scholar * Feng Zhang View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS
F.A.R., P.D.H., J.W., D.A.S. and F.Z. designed and performed the experiments. V.A. contributed to the online tool. F.A.R., P.D.H. and F.Z. wrote the manuscript with help from all authors.
CORRESPONDING AUTHOR Correspondence to Feng Zhang. ETHICS DECLARATIONS COMPETING INTERESTS A patent application has been filed relating to this work. SUPPLEMENTARY INFORMATION SUPPLEMENTARY
DATA 1 Supplementary sequences (PDF 97 kb) SUPPLEMENTARY DATA 2 pSpCas9(BB) plasmid sequence (TXT 12 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE
Ran, F., Hsu, P., Wright, J. _et al._ Genome engineering using the CRISPR-Cas9 system. _Nat Protoc_ 8, 2281–2308 (2013). https://doi.org/10.1038/nprot.2013.143 Download citation * Published:
24 October 2013 * Issue Date: November 2013 * DOI: https://doi.org/10.1038/nprot.2013.143 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get
shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative