Play all audios:
ABSTRACT Ribonucleotides are frequently misincorporated into DNA during replication, and they are rapidly repaired by ribonucleotide excision repair (RER). Although ribonucleotides in
template DNA perturb replicative polymerases and can be considered as DNA damage, they also serve positive biological functions, including directing the orientation of mismatch repair. Here
we describe a method for ribonucleotide identification by high-throughput sequencing that allows mapping of the location of ribonucleotides across the genome. When combined with specific
mutations in the replicative polymerases that incorporate ribonucleotides at elevated frequencies, our ribonucleotide identification method was adapted to map polymerase usage across the
genome. Polymerase usage sequencing (Pu-seq) has been used to define, in unprecedented detail, replication dynamics in yeasts. Although other methods that examine replication dynamics
provide direct measures of replication timing and indirect estimates of origin efficiency, Pu-seq directly ascertains origin efficiency. The Pu-seq protocol can be completed in 12–14 d.
Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this
journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now
Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer
support SIMILAR CONTENT BEING VIEWED BY OTHERS GENOME-WIDE MEASUREMENT OF DNA REPLICATION FORK DIRECTIONALITY AND QUANTIFICATION OF DNA REPLICATION INITIATION AND TERMINATION WITH OKAZAKI
FRAGMENT SEQUENCING Article 18 January 2023 MAPPING RIBONUCLEOTIDES EMBEDDED IN GENOMIC DNA TO SINGLE-NUCLEOTIDE RESOLUTION USING RIBOSE-MAP Article 04 June 2021 MONITORING GENOME-WIDE
REPLICATION FORK DIRECTIONALITY BY OKAZAKI FRAGMENT SEQUENCING IN MAMMALIAN CELLS Article 13 January 2021 REFERENCES * Nick McElhinny, S.A. et al. Abundant ribonucleotide incorporation into
DNA by yeast replicative polymerases. _Proc. Natl. Acad. Sci. USA_ 107, 4949–4954 (2010). Article CAS Google Scholar * Sparks, J.L. et al. RNase H2-initiated ribonucleotide excision
repair. _Mol. Cell_ 47, 980–986 (2012). Article CAS Google Scholar * Lazzaro, F. et al. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. _Mol.
Cell_ 45, 99–110 (2012). Article CAS Google Scholar * Shen, Y., Koh, K.D., Weiss, B. & Storici, F. Mispaired rNMPs in DNA are mutagenic and are targets of mismatch repair and RNases
H. _Nat. Struct. Mol. Biol._ 19, 98–104 (2012). Article CAS Google Scholar * Nick McElhinny, S.A. et al. Genome instability due to ribonucleotide incorporation into DNA. _Nat. Chem.
Biol._ 6, 774–781 (2010). Article CAS Google Scholar * Watt, D.L., Johansson, E., Burgers, P.M. & Kunkel, T.A. Replication of ribonucleotide-containing DNA templates by yeast
replicative polymerases. _DNA Repair (Amst)_ 10, 897–902 (2011). Article CAS Google Scholar * Vengrova, S. & Dalgaard, J.Z. The wild-type _Schizosaccharomyces pombe_ mat1 imprint
consists of two ribonucleotides. _EMBO Rep._ 7, 59–65 (2006). Article CAS Google Scholar * Ghodgaonkar, M.M. et al. Ribonucleotides misincorporated into DNA act as strand-discrimination
signals in eukaryotic mismatch repair. _Mol. Cell_ 50, 323–332 (2013). Article CAS Google Scholar * Lujan, S.A., Williams, J.S., Clausen, A.R., Clark, A.B. & Kunkel, T.A.
Ribonucleotides are signals for mismatch repair of leading-strand replication errors. _Mol. Cell_ 50, 437–443 (2013). Article CAS Google Scholar * Brown, J.A. & Suo, Z. Unlocking the
sugar 'steric gate' of DNA polymerases. _Biochemistry_ 50, 1135–1142 (2011). Article CAS Google Scholar * Reijns, M.A. et al. Lagging-strand replication shapes the mutational
landscape of the genome. _Nature_ 518, 502–506 (2015). Article CAS Google Scholar * Daigaku, Y. et al. A global profile of replicative polymerase usage. _Nat. Struct. Mol. Biol._ 22,
192–198 (2015). Article CAS Google Scholar * Koh, K.D., Balachander, S., Hesselberth, J.R. & Storici, F. Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA. _Nat.
Methods_ 12, 251–257 (2015). Article CAS Google Scholar * Clausen, A.R. et al. Tracking replication enzymology _in vivo_ by genome-wide mapping of ribonucleotide incorporation. _Nat.
Struct. Mol. Biol._ 22, 185–191 (2015). Article CAS Google Scholar * Dellino, G.I. et al. Genome-wide mapping of human DNA-replication origins: levels of transcription at ORC1 sites
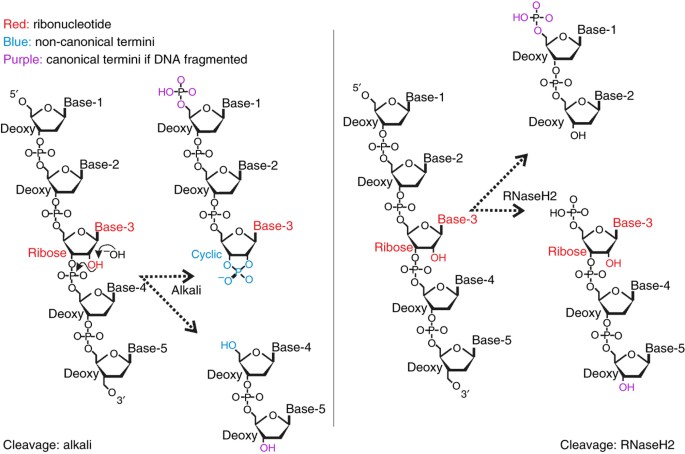
regulate origin selection and replication timing. _Genome Res._ 23, 1–11 (2013). Article CAS Google Scholar * Lipkin, D., Talbert, P.T. & Cohn, M. The mechanism of the alkaline
hydrolysis of ribonucleic acids. _J. Am. Chem. Soc._ 76, 2871–2872 (1954). Article CAS Google Scholar * Koh, K.D., Hesselberth, J.y. & Storici, F. Ribose-seq: ribonucleotides in DNA
to Illumina library. _Protocol Exchange_ 10.1038/protex.2015.044 (2015). * Reijns, M.A. et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and
development. _Cell_ 149, 1008–1022 (2012). Article CAS Google Scholar * Hawkins, M. et al. High-resolution replication profiles define the stochastic nature of genome replication
initiation and termination. _Cell Rep._ 5, 1132–1141 (2013). Article CAS Google Scholar * Retkute, R., Nieduszynski, C.A. & de Moura, A. Mathematical modeling of genome replication.
_Phys. Rev. E Stat. Nonlin. Soft Matter Phys._ 86, 031916 (2012). Article Google Scholar * Moreno, S., Klar, A. & Nurse, P. Molecular genetic analysis of fission yeast
_Schizosaccharomyces pombe_. _Methods Enzymol._ 194, 795–823 (1991). Article CAS Google Scholar * Watson, A.T., Garcia, V., Bone, N., Carr, A.M. & Armstrong, J. Gene tagging and gene
replacement using recombinase-mediated cassette exchange in _Schizosaccharomyces pombe_. _Gene_ 407, 63–74 (2008). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS A.M.C.
acknowledges UK Medical Research Council (MRC) grant no. G1100074 and European Research Council (ERC) grant no. 268788-SMI-DDR. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Genome Damage
and Stability Centre, School of Life Sciences, University of Sussex, Brighton, UK Andrea Keszthelyi, Yasukazu Daigaku, Katie Ptasińska, Izumi Miyabe & Antony M Carr * Frontier Research
Institute for Interdisciplinary Sciences, Tohoku University, Sendai, Japan Yasukazu Daigaku Authors * Andrea Keszthelyi View author publications You can also search for this author inPubMed
Google Scholar * Yasukazu Daigaku View author publications You can also search for this author inPubMed Google Scholar * Katie Ptasińska View author publications You can also search for this
author inPubMed Google Scholar * Izumi Miyabe View author publications You can also search for this author inPubMed Google Scholar * Antony M Carr View author publications You can also
search for this author inPubMed Google Scholar CONTRIBUTIONS A.K., Y.D., K.P. and I.M. performed the experiments. A.K. and Y.D. designed the protocol and analytical methods. A.K., Y.D.,
A.M.C. and K.P. wrote the manuscript. CORRESPONDING AUTHOR Correspondence to Antony M Carr. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests.
RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Keszthelyi, A., Daigaku, Y., Ptasińska, K. _et al._ Mapping ribonucleotides in genomic DNA and exploring
replication dynamics by polymerase usage sequencing (Pu-seq). _Nat Protoc_ 10, 1786–1801 (2015). https://doi.org/10.1038/nprot.2015.116 Download citation * Published: 15 October 2015 * Issue
Date: November 2015 * DOI: https://doi.org/10.1038/nprot.2015.116 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a
shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative