Play all audios:
ABSTRACT High-throughput transcriptional analysis has unveiled a myriad of novel RNAs. However, technical constraints in RNA sequencing library preparation and platform performance hamper
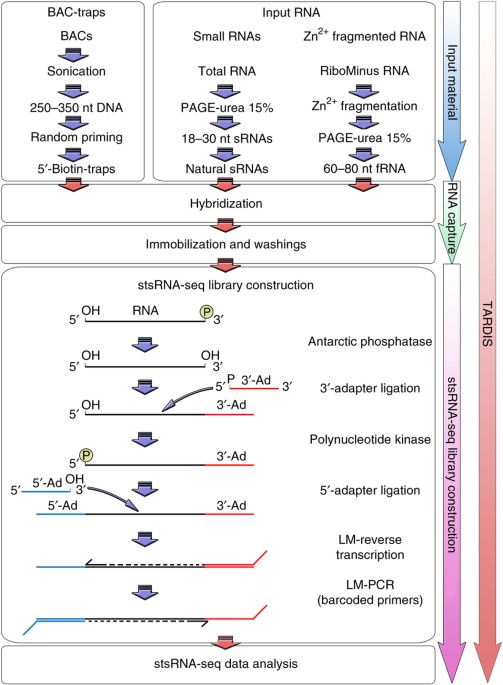
the identification of rare transcripts contained within the RNA repertoire. Herein we present targeted-RNA directional sequencing (TARDIS), a hybridization-based method that allows subsets
of RNAs contained within the transcriptome to be interrogated independently of transcript length, function, the presence or absence of poly-A tracts, or the mechanism of biogenesis. TARDIS
is a modular protocol that is subdivided into four main phases, including the generation of random DNA traps covering the region of interest, purification of input RNA material, DNA
trap–based RNA capture, and finally RNA-sequencing library construction. Importantly, coupling RNA capture to strand-specific RNA sequencing enables robust identification and reconstruction
of novel transcripts, the definition of sense and antisense RNA pairs and, by the concomitant analysis of long and natural small RNA pools, it allows the user to infer potential
precursor-product relations. TARDIS takes ∼10 d to implement. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS
OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on
SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about
institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS OPTIMIZED IDENTIFICATION AND CHARACTERIZATION OF SMALL RNAS WITH PANDORA-SEQ
Article 03 April 2025 NAD TAGSEQ FOR TRANSCRIPTOME-WIDE IDENTIFICATION AND CHARACTERIZATION OF NAD+-CAPPED RNAS Article 03 August 2020 GLOBAL IN SITU PROFILING OF RNA-RNA SPATIAL
INTERACTIONS WITH RIC-SEQ Article 21 May 2021 REFERENCES * Djebali, S. et al. Landscape of transcription in human cells. _Nature_ 489, 101–108 (2012). Article CAS Google Scholar *
Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. _Nat. Methods_ 5, 621–628 (2008). Article CAS Google
Scholar * Trapnell, C. et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. _Nat. Biotechnol._ 28,
511–515 (2010). Article CAS Google Scholar * Hansen, K.D., Brenner, S.E. & Dudoit, S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. _Nucleic Acids
Res._ 38, e131 (2010). Article Google Scholar * Levin, J.Z. et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. _Nat. Methods_ 7, 709–715 (2010). Article
CAS Google Scholar * Cui, P. et al. A comparison between ribo-minus RNA-sequencing and polyA-selected RNA-sequencing. _Genomics_ 96, 259–265 (2010). Article CAS Google Scholar * Aird,
D. et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. _Genome Biol._ 12, R18 (2011). Article CAS Google Scholar * Nakamura, K. et al.
Sequence-specific error profile of Illumina sequencers. _Nucleic Acids Res._ 39, e90 (2011). Article CAS Google Scholar * Benjamini, Y. & Speed, T.P. Summarizing and correcting the GC
content bias in high-throughput sequencing. _Nucleic Acids Res._ 40, e72 (2012). Article CAS Google Scholar * Adiconis, X. et al. Comparative analysis of RNA sequencing methods for
degraded or low-input samples. _Nat. Methods_ 10, 623–629 (2013). Article CAS Google Scholar * Spicuglia, S., Maqbool, M.A., Puthier, D. & Andrau, J.C. An update on recent methods
applied for deciphering the diversity of the noncoding RNA genome structure and function. _Methods_ 63, 3–17 (2013). Article CAS Google Scholar * Lahens, N.F. et al. IVT-seq reveals
extreme bias in RNA sequencing. _Genome Biol._ 15, R86 (2014). Article Google Scholar * Gnirke, A. et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel
targeted sequencing. _Nat. Biotechnol._ 27, 182–189 (2009). Article CAS Google Scholar * Mamanova, L. et al. Target-enrichment strategies for next-generation sequencing. _Nat. Methods_ 7,
111–118 (2010). Article CAS Google Scholar * Mercer, T.R. et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. _Nat. Biotechnol._ 30, 99–104 (2012).
Article CAS Google Scholar * Mercer, T.R. et al. Targeted sequencing for gene discovery and quantification using RNA CaptureSeq. _Nat. Protoc._ 9, 989–1009 (2014). Article CAS Google
Scholar * Portal, M.M., Pavet, V., Erb, C. & Gronemeyer, H. Human cells contain natural double-stranded RNAs with potential regulatory functions. _Nat. Struct. Mol. Biol._ 22, 89–97
(2015). Article CAS Google Scholar * Yin, W., Rossin, A., Clifford, J.L. & Gronemeyer, H. Co-resistance to retinoic acid and TRAIL by insertion mutagenesis into RAM. _Oncogene_ 25,
3735–3744 (2006). Article CAS Google Scholar * Tomlinson, I. et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. _Nat.
Genet._ 39, 984–988 (2007). Article CAS Google Scholar * Kiemeney, L.A. et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. _Nat. Genet._ 40, 1307–1312
(2008). Article CAS Google Scholar * Radtke, I. et al. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. _Proc. Natl. Acad. Sci. USA_ 106, 12944–12949
(2009). Article CAS Google Scholar * Shete, S. et al. Genome-wide association study identifies five susceptibility loci for glioma. _Nat. Genet._ 41, 899–904 (2009). Article CAS Google
Scholar * Nelson, J.W. & Tinoco, I. Jr. Comparison of the kinetics of ribooligonucleotide, deoxyribooligonucleotide, and hybrid oligonucleotide double-strand formation by
temperature-jump kinetics. _Biochemistry_ 21, 5289–5295 (1982). Article CAS Google Scholar * Wang, S., Friedman, A.E. & Kool, E.T. Origins of high sequence selectivity: a stopped-flow
kinetics study of DNA/RNA hybridization by duplex- and triplex-forming oligonucleotides. _Biochemistry_ 34, 9774–9784 (1995). Article CAS Google Scholar * Shelton, V.M. & Morrow,
J.R. Catalytic transesterification and hydrolysis of RNA by zinc(II) complexes. _Inorg. Chem._ 30, 4295–4299 (1991). Article CAS Google Scholar * Wery, M., Descrimes, M., Thermes, C.,
Gautheret, D. & Morillon, A. Zinc-mediated RNA fragmentation allows robust transcript reassembly upon whole transcriptome RNA-seq. _Methods_ 63, 25–31 (2013). Article CAS Google
Scholar * Karolchik, D. et al. The UCSC Table Browser data retrieval tool. _Nucleic Acids Res._ 32, D493–D496 (2004). Article CAS Google Scholar * Kuhn, R.M. et al. The UCSC Genome
Browser Database: update 2009. _Nucleic Acids Res._ 37, D755–D761 (2009). Article CAS Google Scholar * Rhead, B. et al. The UCSC Genome Browser database: update 2010. _Nucleic Acids Res._
38, D613–D619 (2010). Article CAS Google Scholar * External RNA Controls Consortium. Proposed methods for testing and selecting the ERCC external RNA controls. _BMC Genomics_ 6, 150
(2005). * Jiang, L. et al. Synthetic spike-in standards for RNA-seq experiments. _Genome Res._ 21, 1543–1551 (2011). Article CAS Google Scholar * Langmead, B., Trapnell, C., Pop, M. &
Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. _Genome Biol._ 10, R25 (2009). Article Google Scholar * Goecks, J., Nekrutenko, A.,
Taylor, J. & Galaxy, T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. _Genome Biol._ 11, R86
(2010). Article Google Scholar * Martin, J.A. & Wang, Z. Next-generation transcriptome assembly. _Nat. Rev. Genet._ 12, 671–682 (2011). Article CAS Google Scholar * Kim, D. et al.
TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. _Genome Biol._ 14, R36 (2013). Article Google Scholar * Hummel, M., Bonnin, S.,
Lowy, E. & Roma, G. TEQC: an R package for quality control in target capture experiments. _Bioinformatics_ 27, 1316–1317 (2011). Article CAS Google Scholar * Quinlan, A.R. & Hall,
I.M. BEDTools: a flexible suite of utilities for comparing genomic features. _Bioinformatics_ 26, 841–842 (2010). Article CAS Google Scholar * Li, H. et al. The Sequence Alignment/Map
format and SAMtools. _Bioinformatics_ 25, 2078–2079 (2009). Article Google Scholar * Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. _EMBnet J._ 17,
10–12 (2011). Article Google Scholar * Perocchi, F., Xu, Z., Clauder-Munster, S. & Steinmetz, L.M. Antisense artifacts in transcriptome microarray experiments are resolved by
actinomycin D. _Nucleic Acids Res._ 35, e128 (2007). Article Google Scholar Download references ACKNOWLEDGEMENTS We thank M. Philipps, S. Vicaire and B. Jost from the IGBMC Microarray and
Sequencing platform (France Génomique consortium—ANR-10-INBS-0009). M.M.P. and V.P. were supported as postdoctoral fellows of the Ligue Nationale Contre le Cancer. This work was supported by
funds from the Alliance Nationale pour les Sciences de la Vie et de la Santé–Institut Thématique Multi-organismes Cancer–Institut National du Cancer (INCa) grant 'Epigenomics of breast
cancer', EpiPCa, the Ligue National Contre le Cancer (to H.G.; Equipe Labellisée), the INCa and the European Community contract LSHC-CT-2005-518417 'EPITRON'. Support from
the Agence Nationale de la Recherche (ANRT-07-PCVI-0031-01, ANR-10-LABX-0030-INRT and ANR-10-IDEX-0002-02) is also acknowledged. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Institut de
Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Equipe Labellisée Ligue Contre le Cancer, Centre National de la Recherche Scientifique UMR 7104, Institut National de la Santé et
de la Recherche Médicale U964, University of Strasbourg, Illkirch, France Maximiliano M Portal, Valeria Pavet, Cathie Erb & Hinrich Gronemeyer Authors * Maximiliano M Portal View author
publications You can also search for this author inPubMed Google Scholar * Valeria Pavet View author publications You can also search for this author inPubMed Google Scholar * Cathie Erb
View author publications You can also search for this author inPubMed Google Scholar * Hinrich Gronemeyer View author publications You can also search for this author inPubMed Google Scholar
CONTRIBUTIONS M.M.P. conceived and designed the RNA-trap-seq strategy and performed bioinformatics analysis. M.M.P., V.P. and C.E. performed experiments and optimized the final experimental
pipeline. M.M.P., V.P. and H.G. wrote the manuscript. CORRESPONDING AUTHORS Correspondence to Maximiliano M Portal or Hinrich Gronemeyer. ETHICS DECLARATIONS COMPETING INTERESTS A European
patent application, ‘Methods for sequencing and identifying RNAs’ (EP 14 305 822.0) has been filed by M.M.P. and H.G. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE
THIS ARTICLE Portal, M., Pavet, V., Erb, C. _et al._ TARDIS, a targeted RNA directional sequencing method for rare RNA discovery. _Nat Protoc_ 10, 1915–1938 (2015).
https://doi.org/10.1038/nprot.2015.120 Download citation * Published: 29 October 2015 * Issue Date: December 2015 * DOI: https://doi.org/10.1038/nprot.2015.120 SHARE THIS ARTICLE Anyone you
share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the
Springer Nature SharedIt content-sharing initiative