Play all audios:
KEY POINTS * The basic helix–loop–helix transcription factors oligodendrocyte transcription factor 1 (OLIG1) and OLIG2 are structurally related within their DNA targeting domains and, to a
first approximation, are coordinately expressed during development. * Notwithstanding similarities in their protein structure and expression pattern, OLIG1 and OLIG2 have non-overlapping
functions during development and in the postnatal brain. * _Olig2_-null mice have a striking developmental phenotype involving total loss of motor neurons and near-complete loss of
oligodendrocyte progenitors. * The developmental phenotype of _Olig1_-null mice is more nuanced and largely confined to the oligodendrocyte lineage. However, OLIG1 cooperates with OLIG2 in
spinal cord patterning. * A broadening body of literature links OLIG2 to human gliomas, and pathobiological functions of OLIG1 are suggested in the repair of demyelinating injuries. * The
divergent biological and pathobiological functions of OLIG1 and OLIG2 reflect the non-overlapping genetic targets, co-regulator proteins and post-translational modification of these
proteins. ABSTRACT The basic helix–loop–helix transcription factors oligodendrocyte transcription factor 1 (OLIG1) and OLIG2 are structurally similar and, to a first approximation,
coordinately expressed in the developing CNS and postnatal brain. Despite these similarities, it was apparent from early on after their discovery that OLIG1 and OLIG2 have non-overlapping
developmental functions in patterning, neuron subtype specification and the formation of oligodendrocytes. Here, we summarize more recent insights into the separate roles of these
transcription factors in the postnatal brain during repair processes and in neurological disease states, including multiple sclerosis and malignant glioma. We discuss how the unique
functions of OLIG1 and OLIG2 may reflect their distinct genetic targets, co-regulator proteins and/or post-translational modifications. Access through your institution Buy or subscribe This
is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $189.00
per year only $15.75 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated
during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS SINGLE-CELL
TRANSCRIPTOMIC REVEALS MOLECULAR DIVERSITY AND DEVELOPMENTAL HETEROGENEITY OF HUMAN STEM CELL-DERIVED OLIGODENDROCYTE LINEAGE CELLS Article Open access 28 January 2021 SIRT2 PROMOTES WHITE
MATTER OLIGODENDROGENESIS DURING DEVELOPMENT AND IN MODELS OF NEONATAL HYPOXIA Article Open access 15 August 2022 DISTINCT OLIGODENDROCYTE POPULATIONS HAVE SPATIAL PREFERENCE AND DIFFERENT
RESPONSES TO SPINAL CORD INJURY Article Open access 17 November 2020 REFERENCES * Charcot, J. M. Histologie de la sclérose en plaques. _Gazette des hopitaux_ 41, 554–555 (1868) (in French).
Google Scholar * Marburg, O. Die sogenannte akute multiple Sklerose. _Jahrb. Psychiatrie_ 27, 211–312 (1906) (in German). Google Scholar * Prineas, J. W. & Connell, F. Remyelination in
multiple sclerosis. _Ann. Neurol._ 5, 22–31 (1979). Article CAS PubMed Google Scholar * Lucchinetti, C. et al. A quantitative analysis of oligodendrocytes in multiple sclerosis lesions.
A study of 113 cases. _Brain_ 122, 2279–2295 (1999). Article PubMed Google Scholar * Warf, B. C., Fok-Seang, J. & Miller, R. H. Evidence for the ventral origin of oligodendrocyte
precursors in the rat spinal cord. _J. Neurosci._ 11, 2477–2488 (1991). Article CAS PubMed PubMed Central Google Scholar * Pringle, N. P. & Richardson, W. D. A singularity of PDGF
alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. _Development_ 117, 525–533 (1993). CAS PubMed Google Scholar *
Yu, W. P., Collarini, E. J., Pringle, N. P. & Richardson, W. D. Embryonic expression of myelin genes: evidence for a focal source of oligodendrocyte precursors in the ventricular zone of
the neural tube. _Neuron_ 12, 1353–1362 (1994). Article CAS PubMed Google Scholar * Timsit, S. et al. Oligodendrocytes originate in a restricted zone of the embryonic ventral neural
tube defined by DM-20 mRNA expression. _J. Neurosci._ 15, 1012–1024 (1995). Article CAS PubMed PubMed Central Google Scholar * Cameron-Curry, P. & Le Douarin, N. M. Oligodendrocyte
precursors originate from both the dorsal and the ventral parts of the spinal cord. _Neuron_ 15, 1299–1310 (1995). Article CAS PubMed Google Scholar * Raff, M. C., Miller, R. H. &
Noble, M. A glial progenitor cell that develops _in vitro_ into an astrocyte or an oligodendrocyte depending on culture medium. _Nature_ 303, 390–396 (1983). Article CAS PubMed Google
Scholar * Rao, M. S., Noble, M. & Mayer-Proschel, M. A tripotential glial precursor cell is present in the developing spinal cord. _Proc. Natl Acad. Sci. USA_ 95, 3996–4001 (1998).
Article CAS PubMed PubMed Central Google Scholar * Lee, J. C., Mayer-Proschel, M. & Rao, M. S. Gliogenesis in the central nervous system. _Glia_ 30, 105–121 (2000). Article CAS
PubMed Google Scholar * Jessell, T. M. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. _Nature Rev. Genet._ 1, 20–29 (2000). Article CAS PubMed
Google Scholar * Lu, Q. R. et al. Sonic hedgehog-regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. _Neuron_ 25, 317–329 (2000). Article
CAS PubMed Google Scholar * Zhou, Q., Wang, S. & Anderson, D. J. Identification of a novel family of oligodendrocyte lineage-specific basic helix–loop–helix transcription factors.
_Neuron_ 25, 331–343 (2000). Article CAS PubMed Google Scholar * Takebayashi, H. et al. Dynamic expression of basic helix–loop–helix Olig family members: implication of Olig2 in neuron
and oligodendrocyte differentiation and identification of a new member, Olig3. _Mech. Dev._ 99, 143–148 (2000). Article CAS PubMed Google Scholar * Lu, Q. R. et al. Common developmental
requirement for Olig function indicates a motor neuron/oligodendrocyte connection. _Cell_ 109, 75–86 (2002). Article CAS PubMed Google Scholar * Zhou, Q. & Anderson, D. J. The bHLH
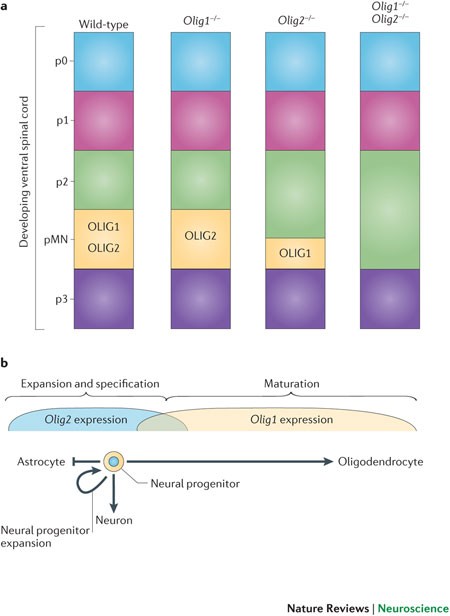
transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. _Cell_ 109, 61–73 (2002). REFERENCES 16–18 COLLECTIVELY SHOW THAT OLIG1 AND OLIG2 REGULATE THE
FORMATION OF OLIGODENDROCYTES AND CERTAIN NEURONS (NOTABLY MOTOR NEURONS) AND HAVE NO APPARENT ROLE IN THE SPECIFICATION OF ASTROCYTES. Article CAS PubMed Google Scholar * Cai, J. et al.
Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. _Neuron_ 45, 41–53 (2005). Article CAS PubMed Google Scholar
* Vallstedt, A., Klos, J. M. & Ericson, J. Multiple dorsoventral origins of oligodendrocyte generation in the spinal cord and hindbrain. _Neuron_ 45, 55–67 (2005). Article CAS PubMed
Google Scholar * Fogarty, M., Richardson, W. D. & Kessaris, N. A subset of oligodendrocytes generated from radial glia in the dorsal spinal cord. _Development_ 132, 1951–1959 (2005).
REFERENCES 19–21 COLLECTIVELY IDENTIFY MULTIPLE ORIGINS OF OLIGODENDROCYTE FORMATION IN THE DEVELOPING CNS. Article CAS PubMed Google Scholar * Menn, B. et al. Origin of oligodendrocytes
in the subventricular zone of the adult brain. _J. Neurosci._ 26, 7907–7918 (2006). Article CAS PubMed PubMed Central Google Scholar * Fancy, S. P., Zhao, C. & Franklin, R. J.
Increased expression of Nkx2.2 and Olig2 identifies reactive oligodendrocyte progenitor cells responding to demyelination in the adult CNS. _Mol. Cell Neurosci._ 27, 247–254 (2004). Article
CAS PubMed Google Scholar * Buffo, A. et al. Expression pattern of the transcription factor Olig2 in response to brain injuries: implications for neuronal repair. _Proc. Natl Acad. Sci.
USA_ 102, 18183–18188 (2005). Article CAS PubMed PubMed Central Google Scholar * Arnett, H. A. et al. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the
CNS. _Science_ 306, 2111–2115 (2004). THIS STUDY DEFINES MYELIN REPAIR FUNCTIONS OF OLIG1 IN MOUSE MODELS OF MS. Article CAS PubMed Google Scholar * Georgieva, L. et al. Convergent
evidence that oligodendrocyte lineage transcription factor 2 (OLIG2) and interacting genes influence susceptibility to schizophrenia. _Proc. Natl Acad. Sci. USA_ 103, 12469–12474 (2006).
Article CAS PubMed PubMed Central Google Scholar * Sims, R. et al. Evidence that variation in the oligodendrocyte lineage transcription factor 2 (_OLIG2_) gene is associated with
psychosis in Alzheimer's disease. _Neurosci. Lett._ 461, 54–59 (2009). Article CAS PubMed Google Scholar * Huang, K. et al. Positive association between OLIG2 and schizophrenia in
the Chinese Han population. _Hum. Genet._ 122, 659–660 (2008). Article CAS PubMed Google Scholar * Chakrabarti, L. et al. _Olig1_ and _Olig2_ triplication causes developmental brain
defects in Down syndrome. _Nature Neurosci._ 13, 927–934 (2010). THIS STUDY SHOWS THAT OLIG1 AND OLIG2 ARE LINKED TO THE BRAIN-SPECIFIC ASPECTS OF DOWN SYNDROME. Article CAS PubMed Google
Scholar * Bouvier, C. et al. Shared oligodendrocyte lineage gene expression in gliomas and oligodendrocyte progenitor cells. _J. Neurosurg._ 99, 344–350 (2003). Article CAS PubMed
Google Scholar * Ligon, K. L. et al. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. _J. Neuropathol. Exp. Neurol._ 63, 499–509 (2004). Article CAS
PubMed Google Scholar * Lu, Q. R. et al. Oligodendrocyte lineage genes (_OLIG_) as molecular markers for human glial brain tumors. _Proc. Natl Acad. Sci. USA_ 98, 10851–10856 (2001).
Article CAS PubMed PubMed Central Google Scholar * Marie, Y. et al. OLIG2 as a specific marker of oligodendroglial tumour cells. _Lancet_ 358, 298–300 (2001). Article CAS PubMed
Google Scholar * Ohnishi, A. et al. Expression of the oligodendroglial lineage-associated markers Olig1 and Olig2 in different types of human gliomas. _J. Neuropathol. Exp. Neurol._ 62,
1052–1059 (2003). REFERENCES 31–34 SHOW THAT EXPRESSION OF GENES FROM THE OLIG FAMILY IS A COMMON FEATURE OF HUMAN GLIOMAS. Article CAS PubMed Google Scholar * Ledent, V., Paquet, O.
& Vervoort, M. Phylogenetic analysis of the human basic helix–loop–helix proteins. _Genome Biol._ 3, research0030–research0030.18 (2002). Article PubMed PubMed Central Google Scholar
* Gray, P. A. et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. _Science_ 306, 2255–2257 (2004). Article CAS PubMed Google Scholar *
Malatesta, P. et al. Neuronal or glial progeny: regional differences in radial glia fate. _Neuron_ 37, 751–764 (2003). Article CAS PubMed Google Scholar * Tsai, H. H. et al. Regional
astrocyte allocation regulates CNS synaptogenesis and repair. _Science_ 337, 358–362 (2012). Article CAS PubMed PubMed Central Google Scholar * Ligon, K. L. et al. Olig2-regulated
lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. _Neuron_ 53, 503–517 (2007). Article CAS PubMed PubMed Central Google Scholar *
Chen, J. A. et al. Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. _Neuron_ 69, 721–735 (2011). Article CAS PubMed PubMed Central
Google Scholar * Takebayashi, H. et al. Non-overlapping expression of Olig3 and Olig2 in the embryonic neural tube. _Mech. Dev._ 113, 169–174 (2002). Article CAS PubMed Google Scholar *
Liu, Z. et al. Control of precerebellar neuron development by Olig3 bHLH transcription factor. _J. Neurosci._ 28, 10124–10133 (2008). Article CAS PubMed PubMed Central Google Scholar *
Lee, S. K., Lee, B., Ruiz, E. C. & Pfaff, S. L. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. _Genes Dev._ 19, 282–294 (2005).
Article CAS PubMed PubMed Central Google Scholar * Li, H., de Faria, J. P., Andrew, P., Nitarska, J. & Richardson, W. D. Phosphorylation regulates OLIG2 cofactor choice and the
motor neuron-oligodendrocyte fate switch. _Neuron_ 69, 918–929 (2011). Article CAS PubMed PubMed Central Google Scholar * Furusho, M. et al. Involvement of the Olig2 transcription
factor in cholinergic neuron development of the basal forebrain. _Dev. Biol._ 293, 348–357 (2006). Article CAS PubMed Google Scholar * Xin, M. et al. Myelinogenesis and axonal
recognition by oligodendrocytes in brain are uncoupled in _Olig1_-null mice. _J. Neurosci._ 25, 1354–1365 (2005). Article CAS PubMed PubMed Central Google Scholar * Marshall, C. A.,
Novitch, B. G. & Goldman, J. E. Olig2 directs astrocyte and oligodendrocyte formation in postnatal subventricular zone cells. _J. Neurosci._ 25, 7289–7298 (2005). Article CAS PubMed
PubMed Central Google Scholar * Cahoy, J. D. et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function.
_J. Neurosci._ 28, 264–278 (2008). CAS PubMed PubMed Central Google Scholar * Chen, Y. et al. The basic helix–loop–helix transcription factor Olig2 is critical for reactive astrocyte
proliferation after cortical injury. _J. Neurosci._ 28, 10983–10989 (2008). Article CAS PubMed PubMed Central Google Scholar * Kageyama, R. & Nakanishi, S. Helix–loop–helix factors
in growth and differentiation of the vertebrate nervous system. _Curr. Opin. Genet. Dev._ 7, 659–665 (1997). Article CAS PubMed Google Scholar * Lee, J. E. Basic helix–loop–helix genes
in neural development. _Curr. Opin. Neurobiol._ 7, 13–20 (1997). Article PubMed Google Scholar * Parras, C. M. et al. Mash1 specifies neurons and oligodendrocytes in the postnatal brain.
_EMBO J._ 23, 4495–4505 (2004). Article CAS PubMed PubMed Central Google Scholar * Singh, S. K. et al. Identification of human brain tumour initiating cells. _Nature_ 432, 396–401
(2004). Article CAS PubMed Google Scholar * Bao, S. et al. Targeting cancer stem cells through L1CAM suppresses glioma growth. _Cancer Res._ 68, 6043–6048 (2008). Article CAS PubMed
PubMed Central Google Scholar * Barrett, L. E. et al. Self-renewal does not predict tumor growth potential in mouse models of high-grade glioma. _Cancer Cell_ 21, 11–24 (2012). Article
CAS PubMed Google Scholar * Appolloni, I. et al. Antagonistic modulation of gliomagenesis by Pax6 and Olig2 in PDGF-induced oligodendroglioma. _Int. J. Cancer_ 131, e1078–e1087 (2012).
Article CAS PubMed Google Scholar * Mehta, S. et al. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and
malignant glioma. _Cancer Cell_ 19, 359–371 (2011). Article CAS PubMed PubMed Central Google Scholar * Kitada, M. & Rowitch, D. H. Transcription factor co-expression patterns
indicate heterogeneity of oligodendroglial subpopulations in adult spinal cord. _Glia_ 54, 35–46 (2006). Article PubMed Google Scholar * Chang, A., Tourtellotte, W. W., Rudick, R. &
Trapp, B. D. Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. _N. Engl. J. Med._ 346, 165–173 (2002). Article PubMed Google Scholar * Kuhlmann, T. et al.
Differentiation block of oligodendroglial progenitor cells as a cause for remyelination failure in chronic multiple sclerosis. _Brain_ 131, 1749–1758 (2008). Article CAS PubMed Google
Scholar * Billiards, S. S. et al. Myelin abnormalities without oligodendrocyte loss in periventricular leukomalacia. _Brain Pathol._ 18, 153–163 (2008). Article PubMed PubMed Central
Google Scholar * Verney, C. et al. Microglial reaction in axonal crossroads is a hallmark of noncystic periventricular white matter injury in very preterm infants. _J. Neuropathol. Exp.
Neurol._ 71, 251–264 (2012). Article CAS PubMed Google Scholar * Fancy, S. P. et al. Axin2 as regulatory and therapeutic target in newborn brain injury and remyelination. _Nature
Neurosci._ 14, 1009–1016 (2011). REFERENCES 59–63 AND 127 COLLECTIVELY SUGGEST THAT DEFECTS IN MYELIN REPAIR IN MS AND PERIVENTRICULAR LEUKOMALACIA REFLECT A FAILURE OF OLIGODENDROCYTE
MATURATION RATHER THAN A LACK OF OLIGODENDROCYTE PROGENITORS. Article CAS PubMed Google Scholar * Pekny, M. & Nilsson, M. Astrocyte activation and reactive gliosis. _Glia_ 50,
427–434 (2005). Article PubMed Google Scholar * Gabay, L., Lowell, S., Rubin, L. L. & Anderson, D. J. Deregulation of dorsoventral patterning by FGF confers trilineage differentiation
capacity on CNS stem cells _in vitro_. _Neuron_ 40, 485–499 (2003). Article CAS PubMed Google Scholar * Setoguchi, T. & Kondo, T. Nuclear export of OLIG2 in neural stem cells is
essential for ciliary neurotrophic factor-induced astrocyte differentiation. _J. Cell Biol._ 166, 963–968 (2004). Article CAS PubMed PubMed Central Google Scholar * Muroyama, Y.,
Fujiwara, Y., Orkin, S. H. & Rowitch, D. H. Specification of astrocytes by bHLH protein SCL in a restricted region of the neural tube. _Nature_ 438, 360–363 (2005). Article CAS PubMed
Google Scholar * Dimou, L., Simon, C., Kirchhoff, F., Takebayashi, H. & Gotz, M. Progeny of Olig2-expressing progenitors in the gray and white matter of the adult mouse cerebral
cortex. _J. Neurosci._ 28, 10434–10442 (2008). Article CAS PubMed PubMed Central Google Scholar * Komitova, M., Serwanski, D. R., Lu, Q. R. & Nishiyama, A. NG2 cells are not a major
source of reactive astrocytes after neocortical stab wound injury. _Glia_ 59, 800–809 (2011). Article PubMed PubMed Central Google Scholar * Zawadzka, M. et al. CNS-resident glial
progenitor/stem cells produce Schwann cells as well as oligodendrocytes during repair of CNS demyelination. _Cell Stem Cell_ 6, 578–590 (2010). Article CAS PubMed Google Scholar *
Tripathi, R. B., Rivers, L. E., Young, K. M., Jamen, F. & Richardson, W. D. NG2 glia generate new oligodendrocytes but few astrocytes in a murine experimental autoimmune
encephalomyelitis model of demyelinating disease. _J. Neurosci._ 30, 16383–16390 (2010). Article CAS PubMed PubMed Central Google Scholar * Buffo, A. et al. Origin and progeny of
reactive gliosis: a source of multipotent cells in the injured brain. _Proc. Natl Acad. Sci. USA_ 105, 3581–3586 (2008). Article CAS PubMed PubMed Central Google Scholar * Magnus, T. et
al. Evidence that nucleocytoplasmic Olig2 translocation mediates brain-injury-induced differentiation of glial precursors to astrocytes. _J. Neurosci. Res._ 85, 2126–2137 (2007). Article
CAS PubMed Google Scholar * Zhao, J. W., Raha-Chowdhury, R., Fawcett, J. W. & Watts, C. Astrocytes and oligodendrocytes can be generated from NG2+ progenitors after acute brain
injury: intracellular localization of oligodendrocyte transcription factor 2 is associated with their fate choice. _Eur. J. Neurosci._ 29, 1853–1869 (2009). Article PubMed Google Scholar
* Cassiani-Ingoni, R. et al. Cytoplasmic translocation of Olig2 in adult glial progenitors marks the generation of reactive astrocytes following autoimmune inflammation. _Exp. Neurol._ 201,
349–358 (2006). Article CAS PubMed Google Scholar * Fernandez, F. & Garner, C. C. Over-inhibition: a model for developmental intellectual disability. _Trends Neurosci._ 30, 497–503
(2007). Article CAS PubMed Google Scholar * Belichenko, P. V. et al. Synaptic structural abnormalities in the Ts65Dn mouse model of Down syndrome. _J. Comp. Neurol._ 480, 281–298 (2004).
Article PubMed Google Scholar * Haydar, T. F. & Reeves, R. H. Trisomy 21 and early brain development. _Trends Neurosciences_ 35, 81–91 (2012). Article CAS Google Scholar * Lu, J.
et al. OLIG2 over-expression impairs proliferation of human Down syndrome neural progenitors. _Hum. Mol. Genet._ 21, 2330–2340 (2012). Article CAS PubMed PubMed Central Google Scholar *
Wang, S. Z. et al. An oligodendrocyte-specific zinc-finger transcription regulator cooperates with Olig2 to promote oligodendrocyte differentiation. _Development_ 133, 3389–3398 (2006).
Article CAS PubMed Google Scholar * Massari, M. E. & Murre, C. Helix–loop–helix proteins: regulators of transcription in eucaryotic organisms. _Mol. Cell. Biol._ 20, 429–440 (2000).
Article CAS PubMed PubMed Central Google Scholar * Zhou, Q., Choi, G. & Anderson, D. J. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration
with Nkx2.2. _Neuron_ 31, 791–807 (2001). Article CAS PubMed Google Scholar * Novitch, B. G., Chen, A. I. & Jessell, T. M. Coordinate regulation of motor neuron subtype identity and
pan-neuronal properties by the bHLH repressor Olig2. _Neuron_ 31, 773–789 (2001). Article CAS PubMed Google Scholar * Mizuguchi, R. et al. Combinatorial roles of olig2 and neurogenin2
in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. _Neuron_ 31, 757–771 (2001). Article CAS PubMed Google Scholar * Kuspert, M., Hammer, A.,
Bosl, M. R. & Wegner, M. Olig2 regulates _Sox10_ expression in oligodendrocyte precursors through an evolutionary conserved distal enhancer. _Nucleic Acids Res._ 39, 1280–1293 (2011).
Article PubMed CAS Google Scholar * Mazzoni, E. O. et al. Embryonic stem cell-based mapping of developmental transcriptional programs. _Nature Methods_ 8, 1056–1058 (2011). Article CAS
PubMed PubMed Central Google Scholar * Weng, Q. et al. Dual-mode modulation of smad signaling by smad-interacting protein sip1 is required for myelination in the central nervous system.
_Neuron_ 73, 713–728 (2012). Article CAS PubMed PubMed Central Google Scholar * Guo, X. et al. Delayed onset of experimental autoimmune encephalomyelitis in Olig1 deficient mice. _PLoS
ONE_ 5, e13083 (2010). Article PubMed PubMed Central CAS Google Scholar * Chen, Y. et al. The oligodendrocyte-specific G protein-coupled receptor GPR17 is a cell-intrinsic timer of
myelination. _Nature Neurosci._ 12, 1398–1406 (2009). Article CAS PubMed Google Scholar * Li, H., Lu, Y., Smith, H. K. & Richardson, W. D. Olig1 and Sox10 interact synergistically to
drive myelin basic protein transcription in oligodendrocytes. _J. Neurosci._ 27, 14375–14382 (2007). Article CAS PubMed PubMed Central Google Scholar * Beckett, D. Regulated assembly
of transcription factors and control of transcription initiation. _J. Mol. Biol._ 314, 335–352 (2001). Article CAS PubMed Google Scholar * Featherstone, M. Coactivators in transcription
initiation: here are your orders. _Curr. Opin. Genet. Dev._ 12, 149–155 (2002). Article CAS PubMed Google Scholar * Torchia, J., Glass, C. & Rosenfeld, M. G. Co-activators and
co-repressors in the integration of transcriptional responses. _Curr. Opin. Cell Biol._ 10, 373–383 (1998). Article CAS PubMed Google Scholar * Ravasi, T. et al. An atlas of
combinatorial transcriptional regulation in mouse and man. _Cell_ 140, 744–752 (2010). Article CAS PubMed Google Scholar * Samanta, J. & Kessler, J. A. Interactions between ID and
OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. _Development_ 131, 4131–4142 (2004). Article CAS PubMed Google Scholar * Benezra, R., Davis, R.
L., Lockshon, D., Turner, D. L. & Weintraub, H. The protein Id: a negative regulator of helix–loop–helix DNA binding proteins. _Cell_ 61, 49–59 (1990). Article CAS PubMed Google
Scholar * Poulin, G., Lebel, M., Chamberland, M., Paradis, F. W. & Drouin, J. Specific protein–protein interaction between basic helix–loop–helix transcription factors and homeoproteins
of the Pitx family. _Mol. Cell. Biol._ 20, 4826–4837 (2000). Article CAS PubMed PubMed Central Google Scholar * Babu, D. A., Chakrabarti, S. K., Garmey, J. C. & Mirmira, R. G. Pdx1
and BETA2/NeuroD1 participate in a transcriptional complex that mediates short-range DNA looping at the insulin gene. _J. Biol. Chem._ 283, 8164–8172 (2008). Article CAS PubMed PubMed
Central Google Scholar * Makarenkova, H. P., Gonzalez, K. N., Kiosses, W. B. & Meech, R. Barx2 controls myoblast fusion and promotes MyoD-mediated activation of the smooth muscle
α-actin gene. _J. Biol. Chem._ 284, 14866–14874 (2009). Article CAS PubMed PubMed Central Google Scholar * Sun, T. et al. Olig bHLH proteins interact with homeodomain proteins to
regulate cell fate acquisition in progenitors of the ventral neural tube. _Curr. Biol._ 11, 1413–1420 (2001). Article CAS PubMed Google Scholar * Fukuda, S., Kondo, T., Takebayashi, H.
& Taga, T. Negative regulatory effect of an oligodendrocytic bHLH factor OLIG2 on the astrocytic differentiation pathway. _Cell Death Differ._ 11, 196–202 (2004). Article CAS PubMed
Google Scholar * Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H. & Nakatani, Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. _Cell_ 87,
953–959 (1996). Article CAS PubMed Google Scholar * Ikushima, H. et al. An Id-like molecule, HHM, is a synexpression group-restricted regulator of TGF-β signalling. _EMBO J._ 27,
2955–2965 (2008). Article CAS PubMed PubMed Central Google Scholar * Huillard, E. et al. Disruption of CK2β in embryonic neural stem cells compromises proliferation and
oligodendrogenesis in the mouse telencephalon. _Mol. Cell. Biol._ 30, 2737–2749 (2010). Article CAS PubMed PubMed Central Google Scholar * Sun, Y. et al. Phosphorylation state of Olig2
regulates proliferation of neural progenitors. _Neuron_ 69, 906–917 (2011). Article CAS PubMed PubMed Central Google Scholar * Sun, T. et al. Cross-repressive interaction of the Olig2
and Nkx2.2 transcription factors in developing neural tube associated with formation of a specific physical complex. _J. Neurosci._ 23, 9547–9556 (2003). Article CAS PubMed PubMed Central
Google Scholar * Niu, J. et al. Phosphorylated olig1 localizes to the cytosol of oligodendrocytes and promotes membrane expansion and maturation. _Glia_ 60, 1427–1436 (2012). Article
PubMed PubMed Central Google Scholar * Gill, G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? _Genes Dev._ 18, 2046–2059 (2004). Article CAS PubMed Google
Scholar * Hay, R. T. SUMO: a history of modification. _Mol. Cell_ 18, 1–12 (2005). Article CAS PubMed Google Scholar * Johnson, E. S. Protein modification by SUMO. _Annu. Rev.
Biochem._ 73, 355–382 (2004). Article CAS PubMed Google Scholar * Verger, A., Perdomo, J. & Crossley, M. Modification with SUMO. A role in transcriptional regulation. _EMBO Rep._ 4,
137–142 (2003). Article CAS PubMed PubMed Central Google Scholar * Huang, T. T., Wuerzberger-Davis, S. M., Wu, Z. H. & Miyamoto, S. Sequential modification of NEMO/IKKγ by SUMO-1
and ubiquitin mediates NF- κB activation by genotoxic stress. _Cell_ 115, 565–576 (2003). Article CAS PubMed Google Scholar * Hay, R. T. Modifying NEMO. _Nature Cell Biol._ 6, 89–91
(2004). Article CAS PubMed Google Scholar * Imoto, S. et al. Sumoylation of Smad3 stimulates its nuclear export during PIASy-mediated suppression of TGF-β signaling. _Biochem. Biophys.
Res. Commun._ 370, 359–365 (2008). Article CAS PubMed Google Scholar * Martin, S., Wilkinson, K. A., Nishimune, A. & Henley, J. M. Emerging extranuclear roles of protein SUMOylation
in neuronal function and dysfunction. _Nature Rev. Neurosci._ 8, 948–959 (2007). Article CAS Google Scholar * Zhou, F., Xue, Y., Lu, H., Chen, G. & Yao, X. A genome-wide analysis of
sumoylation-related biological processes and functions in human nucleus. _FEBS Lett._ 579, 3369–3375 (2005). Article CAS PubMed Google Scholar * Frazer, K. A. et al. Evolutionarily
conserved sequences on human chromosome 21. _Genome Res._ 11, 1651–1659 (2001). Article CAS PubMed PubMed Central Google Scholar * Muskal, S. M., Holbrook, S. R. & Kim, S. H.
Prediction of the disulfide-bonding state of cysteine in proteins. _Protein Eng._ 3, 667–672 (1990). Article CAS PubMed Google Scholar * Smith, E. & Shilatifard, A. The chromatin
signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. _Mol. Cell_ 40, 689–701 (2010). Article CAS PubMed PubMed Central Google
Scholar * el-Husseini Ael, D. & Bredt, D. S. Protein palmitoylation: a regulator of neuronal development and function. _Nature Rev. Neurosci._ 3, 791–802 (2002). Article CAS Google
Scholar * Rowitch, D. H., Lu, Q. R., Kessaris, N. & Richardson, W. D. An. 'oligarchy' rules neural development. _Trends Neurosci._ 25, 417–422 (2002). Article CAS PubMed
Google Scholar * Muller, T. et al. The bHLH factor Olig3 coordinates the specification of dorsal neurons in the spinal cord. _Genes Dev._ 19, 733–743 (2005). Article PubMed PubMed Central
CAS Google Scholar * Ligon, K. L. et al. Development of NG2 neural progenitor cells requires Olig gene function. _Proc. Natl Acad. Sci. USA_ 103, 7853–7858 (2006). Article CAS PubMed
PubMed Central Google Scholar * Miyoshi, G., Butt, S. J., Takebayashi, H. & Fishell, G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic
Olig2-expressing precursors. _J. Neurosci._ 27, 7786–7798 (2007). Article CAS PubMed PubMed Central Google Scholar * Plenge, R. M. et al. Two independent alleles at 6q23 associated with
risk of rheumatoid arthritis. _Nature Genet._ 39, 1477–1482 (2007). Article CAS PubMed Google Scholar * Thomson, W. et al. Rheumatoid arthritis association at 6q23. _Nature Genet._ 39,
1431–1433 (2007). Article CAS PubMed Google Scholar * Buser, J. R. et al. Arrested preoligodendrocyte maturation contributes to myelination failure in premature infants. _Ann. Neurol._
71, 93–109 (2012). Article PubMed PubMed Central Google Scholar * Hack, M. A. et al. Neuronal fate determinants of adult olfactory bulb neurogenesis. _Nature Neurosci._ 8, 865–872
(2005). Article CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS The authors gratefully acknowledge helpful conversations with W. Richardson (University College London,
UK), R. Miller (Case Western Reserve University, USA) and Y. Sun (Dana–Farber Cancer Institute, USA). Work from the authors' laboratories that is cited here was supported by grants from
the National Institutes of Health (NS047572 and NS057727 to C.D.S. and NS040511 to D.H.R.) and from the Pediatric Low-Grade Astrocytoma Foundation (grant awarded to C.D.S.). D.H.R. is
supported by the Howard Hughes Medical Institute. AUTHOR INFORMATION Author notes * Christopher M. Taylor Present address: Present address., AUTHORS AND AFFILIATIONS * Departments of
Neurobiology, Harvard Medical School and Cancer Biology, Dana–Farber Cancer Institute, 450 Brookline Avenue, Boston, 02215, Massachusetts, USA Dimphna H. Meijer, Michael F. Kane, Shwetal
Mehta, Christopher M. Taylor & Charles D. Stiles * Informatics Program, Children's Hospital Boston, 300 Longwood Avenue, Boston, 02115, Massachusetts, USA Hongye Liu * Departments
of Pediatrics and Neurological Surgery, Howard Hughes Medical Institute, University of California, San Francisco, 513 Parnassus Avenue, San Francisco, 94143, California, USA Emily Harrington
& David H. Rowitch * EMD Serono Research Institute, 45A Middlesex Turnpike, Billerica, 01821, Massachusetts, USA Christopher M. Taylor Authors * Dimphna H. Meijer View author
publications You can also search for this author inPubMed Google Scholar * Michael F. Kane View author publications You can also search for this author inPubMed Google Scholar * Shwetal
Mehta View author publications You can also search for this author inPubMed Google Scholar * Hongye Liu View author publications You can also search for this author inPubMed Google Scholar *
Emily Harrington View author publications You can also search for this author inPubMed Google Scholar * Christopher M. Taylor View author publications You can also search for this author
inPubMed Google Scholar * Charles D. Stiles View author publications You can also search for this author inPubMed Google Scholar * David H. Rowitch View author publications You can also
search for this author inPubMed Google Scholar CORRESPONDING AUTHORS Correspondence to Charles D. Stiles or David H. Rowitch. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no
competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION S1 (TABLE) (PDF 161 kb) SUPPLEMENTARY INFORMATION S2 (BOX) (PDF 159 kb) RELATED LINKS RELATED LINKS
DATABASES NCBI Gene expression omnibus database GLOSSARY * Cre-_lox_ fate-mapping This recombination procedure installs a stable marker protein (usually colorimetric, such as LacZ) into a
genetically defined cell type and all of its daughter cells. * Tumour-initiating cells In tumours with a heterogeneous cell population (such as glioblastoma), these undifferentiated
stem-like cells are thought to be responsible for propagating the tumours in serial animal transplantation protocols. * Severe combined immunodeficiency (SCID). SCID mice are used as a host
animal for transplantation experiments with human tumours. * Transit-amplifying cells Also known as type C cells, these are rapidly dividing neural progenitor cells in the subventricular
zone of postnatal brain. They are also the immediate progeny of the more slowly replicating multipotent adult neural stem cells. * Single nucleotide polymorphisms (SNPs). SNPs are DNA
sequence variations that differ among individual members of a biological species or between paired chromosomes in a single individual. * Expression profiling This procedure identifies the
gene types that are expressed in a particular cell type by processing mRNA into cDNA and then annealing the cDNA to gene sequences arrayed onto a solid surface. * SUMOylation This
post-translational modification event involves the covalent ligation of small ubiquitin-like modifier (SUMO) proteins to regulate various cellular processes. RIGHTS AND PERMISSIONS Reprints
and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Meijer, D., Kane, M., Mehta, S. _et al._ Separated at birth? The functional and molecular divergence of OLIG1 and OLIG2. _Nat Rev
Neurosci_ 13, 819–831 (2012). https://doi.org/10.1038/nrn3386 Download citation * Published: 20 November 2012 * Issue Date: December 2012 * DOI: https://doi.org/10.1038/nrn3386 SHARE THIS
ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard
Provided by the Springer Nature SharedIt content-sharing initiative