Play all audios:
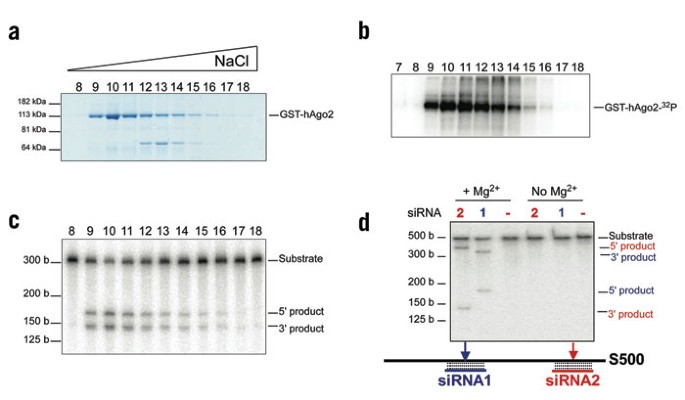
ABSTRACT Genetic, biochemical and structural studies have implicated Argonaute proteins as the catalytic core of the RNAi effector complex, RISC. Here we show that recombinant, human
Argonaute2 can combine with a small interfering RNA (siRNA) to form minimal RISC that accurately cleaves substrate RNAs. Recombinant RISC shows many of the properties of RISC purified from
human or _Drosophila melanogaster_ cells but also has surprising features. It shows no stimulation by ATP, suggesting that factors promoting product release are missing from the recombinant
enzyme. The active site is made up of a unique Asp-Asp-His (DDH) motif. In the RISC reconstitution system, the siRNA 5′ phosphate is important for the stability and the fidelity of the
complex but is not essential for the creation of an active enzyme. These studies demonstrate that Argonaute proteins catalyze mRNA cleavage within RISC and provide a source of recombinant
enzyme for detailed biochemical studies of the RNAi effector complex. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution
ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article *
Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn
about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS MECHANISTIC INSIGHTS INTO RNA CLEAVAGE BY HUMAN ARGONAUTE2–SIRNA COMPLEX
Article 16 April 2025 GTSF1 ACCELERATES TARGET RNA CLEAVAGE BY PIWI-CLADE ARGONAUTE PROTEINS Article Open access 30 June 2022 STRUCTURAL INSIGHTS INTO RNA CLEAVAGE BY PIWI ARGONAUTE Article
15 January 2025 ACCESSION CODES ACCESSIONS GENBANK/EMBL/DDBJ * NP_036286 PROTEIN DATA BANK * 1DI2 * 1G15 * 1MM8 * 1MUR * 1RDD * 1Z25 * 1Z26 REFERENCES * Hannon, G.J. RNA interference.
_Nature_ 418, 244–251 (2002). Article CAS Google Scholar * Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R. & Tuschl, T. Single-stranded antisense siRNAs guide target RNA
cleavage in RNAi. _Cell_ 110, 563–574 (2002). Article CAS Google Scholar * Nykanen, A., Haley, B. & Zamore, P.D. ATP requirements and small interfering RNA structure in the RNA
interference pathway. _Cell_ 107, 309–321 (2001). Article CAS Google Scholar * Lee, Y.S. et al. Distinct roles for _Drosophila_ Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways.
_Cell_ 117, 69–81 (2004). Article CAS Google Scholar * Hammond, S.M., Bernstein, E., Beach, D. & Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in
_Drosophila_ cells. _Nature_ 404, 293–296 (2000). Article CAS Google Scholar * Meister, G. et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. _Mol. Cell_ 15,
185–197 (2004). Article CAS Google Scholar * Liu, J. et al. Argonaute2 is the catalytic engine of mammalian RNAi. _Science_ 305, 1437–1441 (2004). Article CAS Google Scholar * Song,
J.J., Smith, S.K., Hannon, G.J. & Joshua-Tor, L. Crystal structure of Argonaute and its implications for RISC slicer activity. _Science_ 305, 1434–1437 (2004). Article CAS Google
Scholar * Parker, J.S., Roe, S.M. & Barford, D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. _EMBO J._ 23, 4727–4737 (2004).
Article CAS Google Scholar * Martinez, J. & Tuschl, T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. _Genes Dev._ 18, 975–980 (2004). Article CAS Google Scholar *
Schwarz, D.S., Tomari, Y. & Zamore, P.D. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. _Curr. Biol._ 14, 787–791 (2004). Article CAS Google Scholar * Ma, J.B.,
Ye, K. & Patel, D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. _Nature_ 429, 318–322 (2004). Article CAS Google Scholar *
Hockensmith, J.W., Kubasek, W.L., Vorachek, W.R., Evertsz, E.M. & von Hippel, P.H. Laser cross-linking of protein-nucleic acid complexes. _Methods Enzymol._ 208, 211–236 (1991). Article
CAS Google Scholar * Song, J.J. et al. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. _Nat. Struct. Biol._ 10, 1026–1032
(2003). Article CAS Google Scholar * Lingel, A., Simon, B., Izaurralde, E. & Sattler, M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. _Nat. Struct. Mol. Biol._ 11,
576–577 (2004). Article CAS Google Scholar * Tomari, Y., Matranga, C., Haley, B., Martinez, N. & Zamore, P.D. A protein sensor for siRNA asymmetry. _Science_ 306, 1377–1380 (2004).
Article CAS Google Scholar * Elbashir, S.M., Martinez, J., Patkaniowska, A., Lendeckel, W. & Tuschl, T. Functional anatomy of siRNAs for mediating efficient RNAi in _Drosophila
melanogaster_ embryo lysate. _EMBO J._ 20, 6877–6888 (2001). Article CAS Google Scholar * Elbashir, S.M., Lendeckel, W. & Tuschl, T. RNA interference is mediated by 21- and
22-nucleotide RNAs. _Genes Dev._ 15, 188–200 (2001). Article CAS Google Scholar * Stuckey, J.A. & Dixon, J.E. Crystal structure of a phospholipase D family member. _Nat. Struct.
Biol._ 6, 278–284 (1999). Article CAS Google Scholar * Gray, C.H., Good, V.M., Tonks, N.K. & Barford, D. The structure of the cell cycle protein Cdc14 reveals a proline-directed
protein phosphatase. _EMBO J._ 22, 3524–3535 (2003). Article CAS Google Scholar * Cox, D.N. et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem
cell self-renewal. _Genes Dev._ 12, 3715–3727 (1998). Article CAS Google Scholar * Yang, W. & Steitz, T.A. Recombining the structures of HIV integrase, RuvC and RNase H. _Structure_
3, 131–134 (1995). Article CAS Google Scholar * Davies, J.F. 2nd, Hostomska, Z., Hostomsky, Z., Jordan, S.R. & Matthews, D.A. Crystal structure of the ribonuclease H domain of HIV-1
reverse transcriptase. _Science_ 252, 88–95 (1991). Article CAS Google Scholar * Haruki, M., Tsunaka, Y., Morikawa, M., Iwai, S. & Kanaya, S. Catalysis by _Escherichia coli_
ribonuclease HI is facilitated by a phosphate group of the substrate. _Biochemistry_ 39, 13939–13944 (2000). Article CAS Google Scholar * Kanaya, S., Oobatake, M. & Liu, Y. Thermal
stability of _Escherichia coli_ ribonuclease HI and its active site mutants in the presence and absence of the Mg2+ ion. Proposal of a novel catalytic role for Glu48. _J. Biol. Chem._ 271,
32729–32736 (1996). Article CAS Google Scholar * Kanaya, S. & Ikehara, M. Functions and structures of ribonuclease H enzymes. _Subcell. Biochem._ 24, 377–422 (1995). Article CAS
Google Scholar * Steitz, T.A. & Steitz, J.A. A general two-metal-ion mechanism for catalytic RNA. _Proc. Natl. Acad. Sci. USA_ 90, 6498–6502 (1993). Article CAS Google Scholar *
Beese, L.S. & Steitz, T.A. Structural basis for the 3′-5′ exonuclease activity of _Escherichia coli_ DNA polymerase I: a two metal ion mechanism. _EMBO J._ 10, 25–33 (1991). Article CAS
Google Scholar * Goedken, E.R. & Marqusee, S. Co-crystal of _Escherichia coli_ RNase HI with Mn2+ ions reveals two divalent metals bound in the active site. _J. Biol. Chem._ 276,
7266–7271 (2001). Article CAS Google Scholar * Katayanagi, K., Okumura, M. & Morikawa, K. Crystal structure of _Escherichia coli_ RNase HI in complex with Mg2+ at 2.8 Å resolution:
proof for a single Mg(2+)-binding site. _Proteins_ 17, 337–346 (1993). Article CAS Google Scholar * Chapados, B.R. et al. Structural biochemistry of a type 2 RNase H: RNA primer
recognition and removal during DNA replication. _J. Mol. Biol._ 307, 541–556 (2001). Article CAS Google Scholar * Klumpp, K. et al. Two-metal ion mechanism of RNA cleavage by HIV RNase H
and mechanism-based design of selective HIV RNase H inhibitors. _Nucleic Acids Res._ 31, 6852–6859 (2003). Article CAS Google Scholar * Steiniger-White, M., Rayment, I. & Reznikoff,
W.S. Structure/function insights into Tn5 transposition. _Curr. Opin. Struct. Biol._ 14, 50–57 (2004). Article CAS Google Scholar * Davies, D.R., Goryshin, I.Y., Reznikoff, W.S. &
Rayment, I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. _Science_ 289, 77–85 (2000). Article CAS Google Scholar * Steiniger-White, M., Bhasin, A.,
Lovell, S., Rayment, I. & Reznikoff, W.S. Evidence for “unseen” transposase-DNA contacts. _J. Mol. Biol._ 322, 971–982 (2002). Article CAS Google Scholar * Lovell, S., Goryshin, I.Y.,
Reznikoff, W.R. & Rayment, I. Two-metal active site binding of a Tn5 transposase synaptic complex. _Nat. Struct. Biol._ 9, 278–281 (2002). Article CAS Google Scholar * Bujacz, G. et
al. Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity. _J. Biol. Chem._ 272, 18161–18168 (1997). Article CAS
Google Scholar * Peterson, G. & Reznikoff, W. Tn5 transposase active site mutations suggest position of donor backbone DNA in synaptic complex. _J. Biol. Chem._ 278, 1904–1909 (2003).
Article CAS Google Scholar * Haley, B. & Zamore, P.D. Kinetic analysis of the RNAi enzyme complex. _Nat. Struct. Mol. Biol._ 11, 599–606 (2004). Article CAS Google Scholar *
Ishizuka, A., Siomi, M.C. & Siomi, H. A _Drosophila_ fragile X protein interacts with components of RNAi and ribosomal proteins. _Genes Dev._ 16, 2497–2508 (2002). Article CAS Google
Scholar * Tabara, H., Yigit, E., Siomi, H. & Mello, C.C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in _C. elegans_. _Cell_ 109,
861–871 (2002). Article CAS Google Scholar * Tijsterman, M., Ketting, R.F., Okihara, K.L., Sijen, T. & Plasterk, R.H. RNA helicase MUT-14-dependent gene silencing triggered in C.
elegans by short antisense RNAs. _Science_ 295, 694–697 (2002). Article CAS Google Scholar * Tomari, Y. et al. RISC assembly defects in the _Drosophila_ RNAi mutant armitage. _Cell_ 116,
831–841 (2004). Article CAS Google Scholar * Motamedi, M.R. et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. _Cell_ 119, 789–802
(2004). Article CAS Google Scholar * Meister, G. & Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. _Nature_ 431, 343–349 (2004). Article CAS Google Scholar * Haley,
B., Tang, G. & Zamore, P.D. In vitro analysis of RNA interference in _Drosophila melanogaster_. _Methods_ 30, 330–336 (2003). Article CAS Google Scholar * Otwinowski, Z. & Minor,
W. Processing of X-ray diffraction data collected in oscillation mode. _Methods Enzymol._ 276, 307–326 (1997). Article CAS Google Scholar * Jones, T.A. & Kjeldgaard, M.
Electron-density map interpretation. _Methods Enzymol._ 277, 173–208 (1997). Article CAS Google Scholar * Brünger, A.T. et al. Crystallography & NMR system: a new software suite for
macromolecular structure determination. _Acta Crystallogr. D_ 54, 905–921 (1998). Article Google Scholar * Ryter, J.M. & Schultz, S.C. Molecular basis of double-stranded RNA-protein
interactions: structure of a dsRNA-binding domain complexed with dsRNA. _EMBO J_ 17, 7505–7513 (1998). Article CAS Google Scholar * Esnouf, R.M. An extensively modified version of
MolScript that includes greatly enhanced coloring capabilities. _J. Mol. Graph._ 15, 132–134 (1997). Article CAS Google Scholar * Bacon, D.J. & Anderson, W.F. A fast algorithm for
rendering space-filling molecule pictures. _J. Mol. Graph._ 6, 219–220 (1988). Article Google Scholar * Merritt, E.A. & Murphy, M.E.P. Raster3D version 2.0—a program for photorealistic
molecular graphics. _Acta Crystallogr. D_ 50, 869–873 (1994). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We thank members of the Hannon and Joshua-Tor laboratories
for helpful discussions and A. Heroux (X26C) for support with data collection at the National Synchrotron Light Source (NSLS). C. Marsden and S. Smith provided technical support. P. Zamore
kindly provided kinetic tutoring. The NSLS is supported by the US Department of Energy, Division of Material Sciences and Division of Chemical Sciences. F.V.R. is a fellow of the Jane Coffin
Childs Memorial Fund. J.J.S. is a Bristol-Myers Squibb Predoctoral Fellow. This work was supported in part by a grant from the US National Institutes of Health (G.J.H.) and the Louis Morin
Charitable Trust (L.J.). AUTHOR INFORMATION Author notes * Fabiola V Rivas, Niraj H Tolia and Ji-Joon Song: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * Cold
Spring Harbor Laboratory, Watson School of Biological Sciences, 1 Bungtown Road, Cold Spring Harbor, 11724, New York, USA Fabiola V Rivas, Niraj H Tolia, Ji-Joon Song, Juan P Aragon, Jidong
Liu, Gregory J Hannon & Leemor Joshua-Tor * W.M. Keck Structural Biology Laboratory, 1 Bungtown Road, Cold Spring Harbor, 11724, New York, USA Niraj H Tolia, Ji-Joon Song & Leemor
Joshua-Tor Authors * Fabiola V Rivas View author publications You can also search for this author inPubMed Google Scholar * Niraj H Tolia View author publications You can also search for
this author inPubMed Google Scholar * Ji-Joon Song View author publications You can also search for this author inPubMed Google Scholar * Juan P Aragon View author publications You can also
search for this author inPubMed Google Scholar * Jidong Liu View author publications You can also search for this author inPubMed Google Scholar * Gregory J Hannon View author publications
You can also search for this author inPubMed Google Scholar * Leemor Joshua-Tor View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHORS
Correspondence to Gregory J Hannon or Leemor Joshua-Tor. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY
FIG. 1 Slicer activity is intrinsic to recombinant Ago2. (PDF 3987 kb) SUPPLEMENTARY FIG. 2 The 5′ phosphate contributes to the formation of active RISC. (PDF 704 kb) SUPPLEMENTARY FIG. 3
Tungstate-binding sites. (PDF 628 kb) SUPPLEMENTARY FIG. 4 Mn2+-bound PfAgo. (PDF 561 kb) SUPPLEMENTARY FIG. 5 ATP does not accelerate cleavage by recombinant RISC. (PDF 595 kb)
SUPPLEMENTARY FIG. 6 Kinetic analysis of recombinant RISC. (PDF 917 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Rivas, F., Tolia, N., Song, JJ.
_et al._ Purified Argonaute2 and an siRNA form recombinant human RISC. _Nat Struct Mol Biol_ 12, 340–349 (2005). https://doi.org/10.1038/nsmb918 Download citation * Received: 22 December
2004 * Accepted: 09 March 2005 * Published: 30 March 2005 * Issue Date: 01 April 2005 * DOI: https://doi.org/10.1038/nsmb918 SHARE THIS ARTICLE Anyone you share the following link with will
be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt
content-sharing initiative