Play all audios:
ABSTRACT Recent studies have focused on identifying novel genes involved in the browning process of inguinal white adipose tissue (iWAT). In this context, we propose that the mitochondrial
ATPase gene lactation elevated 1 (Lace1) utilizes lactate to regulate the browning capacity of iWAT, specifically in response to challenge with CL-316,243 (CL), a beta3-adrenergic receptor
(β3-AR) agonist. The mice were injected with CL over a span of 3 days and exposed to cold temperatures (4–6 °C) for 1 week. The results revealed a significant increase in Lace1 expression
levels during beige adipogenesis. Additionally, a strong positive correlation was observed between _Lace1_ and _Ucp1_ mRNA expression in iWAT under browning stimulation. To further explore
this phenomenon, we subjected engineered Lace1 KO mice to CL and cold challenges to validate their browning potential. Surprisingly, Lace1 KO mice presented increased oxygen consumption and
heat generation upon CL challenge and cold exposure, along with increased expression of genes related to brown adipogenesis. Notably, deletion of Lace1 led to increased lactate uptake and
browning in iWAT under CL challenge compared with those of the controls. These unique phenomena stem from increased lactate release due to the inactivation of pyruvate dehydrogenase (PDH) in
the hearts of Lace1 KO mice. SIMILAR CONTENT BEING VIEWED BY OTHERS _FAS_ MUTATION REDUCES OBESITY BY INCREASING IL-4 AND IL-10 EXPRESSION AND PROMOTING WHITE ADIPOSE TISSUE BROWNING
Article Open access 20 July 2020 _EGR1_ LOSS-OF-FUNCTION PROMOTES BEIGE ADIPOCYTE DIFFERENTIATION AND ACTIVATION SPECIFICALLY IN INGUINAL SUBCUTANEOUS WHITE ADIPOSE TISSUE Article Open
access 28 September 2020 PERILIPIN 5 LINKS MITOCHONDRIAL UNCOUPLED RESPIRATION IN BROWN FAT TO HEALTHY WHITE FAT REMODELING AND SYSTEMIC GLUCOSE TOLERANCE Article Open access 03 June 2021
INTRODUCTION Stimuli such as cold exposure, treatment with β3-adrenergic receptor (β3-AR) agonizts such as CL-316,243 (CL), and exercise can induce the conversion of inguinal white adipose
tissue (iWAT) into beige adipose tissue1. This phenomenon is referred to as the “browning” of white adipocytes2. Beige adipocytes are similar to brown adipocytes and present characteristics
such as multilocular lipid droplets and abundant mitochondria. Their primary role involves the dissipation of energy to generate heat. Therefore, the browning of iWAT is recognized as an
important mechanism in obesity research. CL, an agonist of β3-AR, stimulates β3-AR by binding to its G protein α-subunit. This activation initiates the protein kinase A (PKA) pathway within
adipocytes by triggering adenylate cyclase and cyclic AMP (cAMP) production3. Consequently, this process induces lipolysis, which converts triacylglycerol (TG) into fatty acids (FAs) via the
phosphorylation of hormone-sensitive lipase (HSL) in adipocytes. The generated free FAs then activate the induction of uncoupling protein 1 (Ucp1) in mitochondria, which is the thermogenic
pathway4. UCP1 is a mitochondrial protein and a key gene that is upregulated during the browning process5. Recent studies have revealed novel forms of Ucp1-independent thermogenesis, but
these processes have been poorly explored6,7,8. These current research trends have ignited the quest to identify new genes involved in the browning process. Hence, we conducted an RNA-seq
analysis of iWAT following CL treatment. The aim of this study was to assess the genes whose expression is upregulated in beige adipocytes compared with white adipocytes. From the pool of
292 genes whose expression was upregulated in the CL-injected group, 97 mitochondrial genes were selected for further investigation. Within this subset, we identified 11 genes that were
enriched in BAT, leveraging the BioGPS database. Among these candidates, we chose to focus on lactation elevated 1 (Lace1), a gene with an as-yet-undetermined function and role, for our
study. Lace1 is a human homolog of the yeast mitochondrial ATPase family gene 1 (Afg1), a member of the SEC18-NSF, PAS1, CDC48-VCP, and TBP families9. ATPases are categorized into F-ATPase,
P-ATPase, and V-ATPase, of which Lace1 is classified as an F0/F1 ATPase10. F0/F1 ATP synthase is an enzyme localized in the inner membrane of mitochondria, where it catalyzes the synthesis
of ATP from ADP11. Recent studies have reported that ATPases regulate thermogenesis in a Ucp1-dependent or Ucp1-independent manner12,13. Lace1 is also a mitochondrial integral membrane
protein that controls mitochondrial protein homeostasis9. Cesnekova et al. reported that Lace1 mediates the degradation of the nuclear-encoded complex IV subunits cytochrome c oxidase 4
(COX4), COX5A, and COX6A and is required for the normal activity of complexes III and IV of the respiratory chain9. Consequently, Lace1, identified as a mitochondrial gene, has increased
expression due to CL or cold exposure. These observations strongly suggest that Lace1 might serve as a potential novel marker for browning. These findings prompted us to further explore the
role of Lace1 in white fat browning and ascertain its importance. Our approach involved the generation of Lace1 knockout (KO) mice. Our findings revealed that the Lace1 gene was upregulated
during the browning process of iWAT. Strikingly, Lace1 KO mice displayed an increased capacity for browning. This intriguing outcome prompted us to meticulously document and analyze the
intricate mechanisms behind this phenomenon. In this study, we investigated the role of Lace1 in white adipose tissue browning. MATERIALS AND METHODS ANIMALS AND EXPERIMENTAL DESIGN All
animal experiments were performed according to the “Guide for Animal Experiments” (edited by the Korean Academy of Medical Sciences). These animal experiments were approved by the
Institutional Animal Care and Use Committee of Seoul National University, Seoul, Korea (permission number: SNU-210217-9). Lace1 KO mice were used to delete exon 2 of the Lace1 gene via the
CRISPR-Cas9 system by Macrogen (Korea). All the animals were housed under a 12-h light/dark cycle at 22 ± 1 °C and 50–60% humidity with a 12 h light/dark cycle in a specific pathogen-free
facility. PHYSIOLOGICAL MEASUREMENT The fat mass and lean mass of the mice were measured via nuclear magnetic resonance (NMR) methods (Minispec LF-50, Bruker, Germany). The rectal
temperature of the mice was recorded via a BAT-12 microprobe thermometer (Physitemp, USA). PROTEIN EXTRACTION AND WESTERN BLOTTING For protein expression analysis, all proteins were
extracted via RIPA buffer (BR002, Biosolution, Korea) supplemented with protease inhibitor cocktail and phosphatase inhibitor cocktail (P3100-001, P3200-001, GeneDEPOT, USA). Equal amounts
of proteins were separated on SDS‒PAGE gels and transferred to PVDF membranes. Primary antibodies targeting the following proteins were used: LACE1 (NBP1-89215, Novus Biologicals, CO, USA),
UCP1 (ab10983, Abcam), MCT1 (ab250131, Abcam), MCT4 (ab74109, Abcam), LDHA (2012, CST), phospho-PDH (31866, CST), PDH (3205, CST), α-actin (A2066, Sigma‒Aldrich), and β-tubulin (ab2146,
Abcam). RNA EXTRACTION AND QUANTITATIVE REAL-TIME PCR For mRNA expression analysis, total RNA was extracted via TRIzol reagent (Invitrogen), and 1 µg of RNA was synthesized into cDNA via RT
premix (K-2044-B, Bioneer, Korea). qPCR was performed via the SYBR Green Kit (BIO-92005, Meridian Bioscience, OH, USA) according to the manufacturer’s instructions. The fold change for all
the samples was calculated via the 2−ΔΔCt method. The primer sequences are listed in Table 1. HISTOPATHOLOGY All the tissues were fixed with 4% paraformaldehyde (HP2031, Biosesang) at room
temperature overnight, embedded in paraffin, and sectioned into 3–5 μm sections. For H&E staining, all the slides were stained following the manufacturer’s instructions. For
immunohistochemical staining, the slides were stained with an anti-UCP1 antibody (ab10983, Abcam). INDIRECT CALORIMETRY VO2 and energy expenditure were estimated by indirect calorimetry
during CL challenge for 3 days and cold exposure (4 °C) for 7 days. During indirect calorimetry, all the mice were housed in a single cage with free access to food and water under a 12 h
light/dark cycle. All the mice were monitored by PhenoMaster 7.5.6 (TSE system, Germany) during the measurement of VO2 and energy expenditure. L-LACTATE MEASUREMENT Serum L-lactate levels
were measured with an L-lactate assay kit (ab65331, Abcam, UK). All protocols were performed following the manufacturer’s instructions. ANALYSIS OF BULK-RNA SEQUENCING AND BIOINFORMATICS
DATA lllumina’s TruSeq Stranded mRNA LT Sample Prep Kit was used to prepare RNA-sequencing libraries and high-throughput sequencing was performed with Illumina’s NovaSeq 6000 Platform for
each sample. The sequenced reads were mapped to the GRCm39 mouse reference genome via STAR v2.7.4a. Differential expression analysis was performed via the R packages DESeq2 v.1.32.0 and
apeglm v.1.14.0 via the shrinkage method. Differentially expressed genes were identified with cutoffs of adjusted _P_ values < 0.01 and log2-fold changes > 1 in the CL-challenged
experimental group and adjusted _P_ values < 0.05 and log2-fold changes > 0.58 in the Lace1 KO experimental group. All graphical visualizations were implemented in R via ggplot2 v3.3.5
BROWN ADIPOCYTE CULTURE When the brown preadipocyte cells reached 90–100% confluency, the cells were induced with induction medium (DMEM with 10% FBS, 0.5 mM isobutylmethylxanthine (I7018,
Sigma, USA), 0.5 μM dexamethasone (D1756, Sigma), 125 μM indomethacin (I7378, Sigma), 1 nM T3 (T2877, Sigma) and 20 nM insulin (sc-360248, Santa Cruz)). After induction for 2 days, the
medium was changed to insulin medium (DMEM with 10% FBS, 1 nM T3 and 20 nM insulin). Five days after differentiation, brown adipocytes were transfected with 50 nM Lace1 siRNA (30 nM) or
negative control siRNA for 36 h via Lipofectamine RNAiMAX reagent (13778075, Thermo Scientific, USA) to generate Lace1-knockdown adipocytes. The predesigned siRNA directed against murine
Lace1 mRNA (accession no. NM_001359297.1) was purchased from Bioneer (Daejeon, Korea). BEIGE ADIPOCYTE CULTURE Seven-week-old male C57BL/6N mice were sacrificed, and the stromal vascular
fraction (SVF) was harvested from iWAT. The tissues were digested with 1.5 U/ml collagenase D (11088882001, Roche) in a 37 °C shaker for 30 min. The SVF pellets were incubated in
collagen-coated plates with maintenance medium (DMEM/F12 with 10% FBS and 1X P/S) for 1 hr. After that, the medium was changed to fresh medium for removing immune cells, etc. Beige
adipogenesis of preadipocytes of iWAT was induced according to a previous protocol14,15. MITOCHONDRIAL ISOLATION Mitochondrial and cytosolic isolation from differentiated brown adipocytes
was performed with a mitochondrial isolation kit (ab110170, Abcam). All protocols were performed following the manufacturer’s instructions. ATP ASSAY ATP levels in fully differentiated brown
adipocytes were analyzed via an ATP assay kit (K354-100, BioVision, CA, USA). All protocols were performed following the manufacturer’s instructions. OXYGEN CONSUMPTION RATIO (OCR) The
oxygen consumption rate (OCR) of differentiated brown adipocytes from the control siRNA and _siLace1_ knockdown (KD) groups was measured via a Seahorse Fe Extracellular Flux Analyzer
(Agilent Technologies, CA, USA). For the measurement of the OCR, differentiated brown adipocytes were incubated at 37 °C in a non-CO2 incubator for 1 h with Seahorse XF DMEM (103575–100,
Agilent) supplemented with 1 mM pyruvate (103577–100, Agilent), 2 mM glutamine (103579–100, Agilent), and 10 mM glucose (103578–100, Agilent). After incubation, the brown adipocytes were
treated with 1.5 μM oligomycin, 2.0 μM carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP), and 0.5 µM rotenone and antimycin (Rot/AA). All reagents used were from the Agilent
Seahorse XF Cell Mito Stress Test Kit (103015-100, Agilent). The OCR of brown adipocytes was normalized to the protein amount. STATISTICAL ANALYSIS All the statistical data were analyzed via
GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA, USA). All the data are presented as the means ± standard errors of the means (SEMs). Statistical significance was analyzed by an
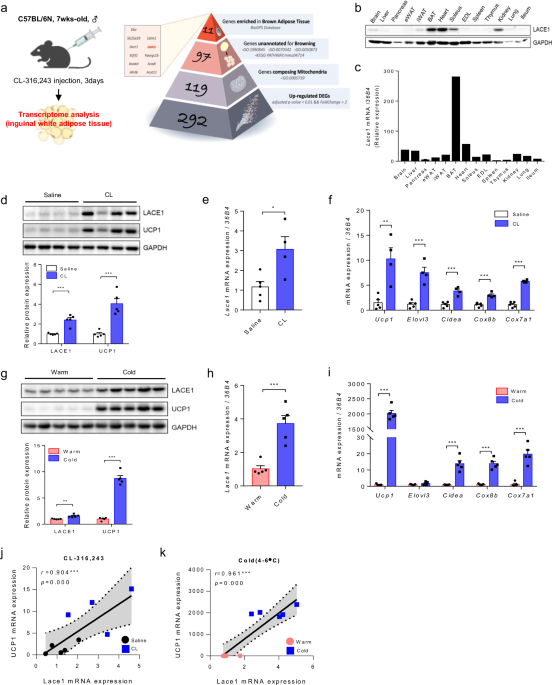
unpaired _t-_test. Statistical significance was determined at *_p_ < 0.05, **_p_ < 0.01 and ***_p_ < 0.001. RESULTS LACE1 IS A MITOCHONDRIAL ATPASE THAT IS HIGHLY EXPRESSED IN BAT
To investigate novel genes involved in beige adipogenesis, we compared the CL-treated and untreated mice via the bulk RNA-seq technique. Among the 292 genes upregulated by CL challenge
(adjusted _P_ value < 0.01 and a fold change > 2 as the cutoff), 97 genes were identified as nonbrowning mitochondrial genes; these genes were annotated as mitochondrial genes
(GO:0005739) and unannotated for browning-related biological processes (GO:1990845, GO:0070342, GO:0050873, KEGG PATHWAY: mmu04714). Using the BioGPS database to identify candidate genes of
interest, we selected 11 genes highly expressed in BAT. In this research, we present Lace1, a highly enriched nonbrowning mitochondrial-annotated gene in BAT, as a novel gene involved in
beige adipogenesis (Fig. 1a and Supplementary Fig. 3). To clarify the involvement of this novel gene, Lace1, in beige adipogenesis, we first measured the expression level of Lace1 in the
whole tissues of 7-week-old C57BL/6N male mice. The expression of LACE1 protein was highest in BAT, followed by the heart, soleus, and kidney (Fig. 1b). Compared with that in other tissues,
_Lace1_ mRNA was highly expressed in BAT (Fig. 1c). Next, as Lace1 expression is highest in BAT, we measured Lace1 expression during brown adipogenesis in immortalized brown preadipocytes.
As a result, LACE1 protein levels were found to be increased during brown adipogenesis, similar to those of Ucp1 and peroxisome proliferator-activated receptor gamma coactivator 1-alpha
(Pgc1α) (Supplementary Fig. 1a). _Lace1_ mRNA was also increased during brown adipogenesis in a _Ucp1-_, cell death-inducing DNA fragmentation factor alpha-like effector A _(Cidea)_- and
_Pgc1α_-dependent manner (Supplementary Fig. 1b–e). During beige adipogenesis, LACE1 protein levels, similar to those of UCP1, increased (Supplementary Fig. 1f). Additionally, _Lace1_ mRNA
expression increased during beige adipogenesis in a _Ucp1_ mRNA expression-dependent manner (Supplementary Fig. 1g, h). These data suggest that Lace1 is highly expressed in the BAT of mice
and that its expression increases during brown adipogenesis and beige adipogenesis. To determine the localization of Lace1 expression, we confirmed the expression of Lace1 in the cytosol and
mitochondria of preadipocytes and fully differentiated brown adipocytes via immortalized brown preadipocytes15. As expected, the LACE1 protein was expressed only in the mitochondria of
fully differentiated brown adipocytes (Supplementary Fig. 2a). Consistent with these findings, we transfected fully differentiated brown adipocytes with siRNAs targeting the Lace1 gene.
After transfection with _siLace1_, we attempted to confirm mitochondrial function in fully differentiated brown adipocytes by directly measuring the oxygen consumption rate (OCR). We found
that the OCR of the _Lace1_-knockdown (KD) group of brown adipocytes was lower than that of the control siRNA group (Supplementary Fig. 2b–f). In particular, basal respiration and maximal
respiration were lower in the Lace1-KD brown adipocytes than in the WT brown adipocytes. Lace1 is also known as a mitochondrial ATPase, which synthesizes ATP9,16. As with previous studies,
we confirmed that ATP levels were lower in the _Lace1_-KD brown adipocytes than in the WT brown adipocytes (Supplementary Fig. 2g). LACE1 IS INCREASED IN IWAT FOLLOWING CL CHALLENGE AND COLD
EXPOSURE Our transcriptomic data revealed that the _Lace1_ gene is one of the genes upregulated in iWAT by CL challenge. Therefore, we hypothesized that Lace1 is increased in beige fat in a
Ucp1 expression-dependent manner and is upregulated during beige adipogenesis. As expected, we confirmed that LACE1 protein levels, similar to those of Ucp1, are increased in CL-induced
beige fat (Fig. 1d). Additionally, _Lace1_ mRNA levels increased, similar to those of thermogenic genes (Fig. 1e and f). Next, we investigated Lace1 expression under cold exposure. Cold
exposure induces browning of iWAT via the release of NE and β3-adrenergic stimulation17,18. Consequently, LACE1 protein levels were greater in cold-induced beige fat than in white fat (Fig.
1g). Similarly, _Lace1_ mRNA expression increased, similar to that of thermogenic genes (Fig. 1h and i). We found that Lace1 was increased in a Ucp1-dependent manner in response to CL
challenge and cold exposure; therefore, we performed a correlation analysis between _Lace1_ and _Ucp1_ mRNA expression. Interestingly, _Lace1_ and _Ucp1_ mRNA levels were positively
correlated under CL challenge and cold exposure (CL challenge: _r_ = 0.904, _p_ = 0.000/cold exposure: _r_ = 0.961, _p_ = 0.000) (Fig. 1j and k). These data suggested that Lace1 is increased
in iWAT following CL challenge and cold exposure. LACE1 DEFICIENCY INCREASES BROWNING OF IWAT FOLLOWING CL INJECTION To investigate the role of Lace1 in beige fat, we generated Lace1 null
knockout (KO) mice via the CRISPR-CAS9 system. A scheme of the construction of Lace1 KO mice is presented in Supplementary Fig. 4. We administered CL to Lace1 KO mice according to this
experimental design (Fig. 2a). First, we confirmed that _Lace1_ protein and mRNA expression is downregulated in the iWAT of the Lace1 KO mice compared with the WT mice despite CL injection
(Figs. 2b and c). Next, we investigated the browning capacity of iWAT in the Lace1 KO mice under CL challenge. Surprisingly, histological analysis revealed that the Lace1 KO mice had a
greater beige fat content than the WT mice when challenged with CL (Fig. 2d). Together, the results of the UCP1 immunostaining analysis revealed that the Lace1 KO mice had more Ucp1-positive
adipocytes than the WT mice did under CL challenge (Fig. 2e). Additionally, thermogenic marker gene levels were greater in the CL-induced beige fat of the Lace1 KO mice than in that of
their control littermates (Fig. 2f). Ucp1 is critical for maintaining core temperature and increasing energy expenditure19. CL challenge and cold exposure-dependent oxygen consumption rate
(VO2) are known to be derived from Ucp1 activity20. To explore the systemic metabolic effects of heightened browning in white adipose tissue of Lace1 KO mice, we conducted indirect
calorimetry during the CL challenge. The Lace1 KO mice exhibited increased oxygen consumption (Fig. 2g) and heat generation after CL challenge (Fig. 2h). We hypothesized that the browning
capacity of iWAT is decreased by Lace1 gene deletion compared with that of WT iWAT because Lace1 is upregulated in iWAT under browning stimuli such as Ucp1; however, surprisingly, we found
that Lace1 deficiency increases the browning capacity of iWAT under CL challenge. On the basis of the above results, we explored whether this phenotype occurred in the Lace1 KO mice upon
exposure to another stimulus, cold, as with the CL challenge. LACE1 DEFICIENCY RESULTS IN INCREASED BROWNING OF IWAT FOLLOWING COLD EXPOSURE For analysis of the phenotype of Lace1 KO mice in
response to cold stimuli, the mice were exposed to cold at 4–6 °C for 7 days (Fig. 3a). As shown in Fig. 3b and c, _Lace1_ protein and mRNA expression was downregulated in the iWAT of the
Lace1 KO mice compared with that of the WT mice despite cold exposure. Histological analysis revealed that the Lace1 KO mice had greater beige fat content than the WT mice when challenged
with cold (Fig. 3d). Additionally, Ucp1 immunostaining analysis revealed that the iWAT of the Lace1 KO mice had more Ucp1-positive adipocytes than did that of the WT mice upon cold exposure
(Fig. 3e). Additionally, thermogenesis-related gene expression was greater in the iWAT of the Lace1 KO mice than in that of the WT mice (Fig. 3f). Compared with those of WT mice, we
subsequently analyzed the systemic metabolic characteristics of Lace1 KO mice after cold exposure and observed significant increases in both VO2 (Fig. 3g) and thermogenesis (Fig. 3h).
Furthermore, we found that the Lace1 KO mice have higher rectal temperatures than the WT mice under cold conditions (Fig. 3i). These findings indicate that the Lace1 KO mice exhibit elevated
energy expenditure when subjected to cold exposure, demonstrating an increased cold tolerance and capacity to sustain core body temperature through heightened thermogenesis. This augmented
thermogenic response strongly aligns with the histological and molecular results, confirming that deficiency of the Lace1 gene promotes browning of iWAT. LACE1 LOSS PROMOTES LACTATE/GPR81
SIGNALING IN CL-INDUCED IWAT To investigate why Lace1 deficiency accelerates the browning of iWAT, we compared the genes whose expression was upregulated in iWAT between the WT and Lace1 KO
mice subjected to CL challenge via bulk RNA sequencing (Supplementary Fig. 5). The RNA-seq data showed that hydroxycarboxylic acid receptor 1 (_Hcar1_) was upregulated in the iWAT of the
Lace1 KO mice upon CL compared with that in the wild-type mice was (Fig. 4a). Hcar1 is a well-known G protein-coupled receptor gene that functions as a receptor for lactate21,22. Hcar1, also
known as Gpr81, is expressed in adipose tissue, liver, and muscle, and its upregulation in iWAT has been reported in response to CL challenge23. On the basis of our sequencing results and
literature, we hypothesized that the Lace1 KO mice exhibit greater browning capacity than the WT mice through lactate utilization. Lactate, a well-known promoter of browning, serves as the
foundation for this hypothesis24. Initially, we quantified the serum L-lactate levels in response to CL treatment. Intriguingly, we observed an elevated amount of circulating L-lactate in
the CL-treated Lace1 KO mice compared with the WT mice (Fig. 4b). We proceeded to assess the expression levels of monocarboxylate transporter 1 (Mct1), which is responsible for lactate
influx, to gauge the lactate uptake capacity in the iWAT of the Lace1 KO mice. First, we analyzed _Mct1_ mRNA levels in the entire tissues of the CL-treated Lace1 KO mice. Notably, _Mct1_
mRNA expression increased more than 10-fold exclusively in the iWAT of the Lace1 KO mice upon CL treatment, with no concurrent alterations in other tissues (Fig. 4c). Additionally, we
observed a substantial increase in MCT1 protein levels, specifically in the iWAT of the Lace1 KO mice subjected to CL treatment (Fig. 4d). Conversely, the expression of the lactate efflux
transporter monocarboxylate transporter 4 (Mct4) remained unaltered in the iWAT of the Lace1 KO mice subjected to CL challenge (Fig. 4e). These findings collectively indicate that, in
comparison with their control counterparts, the Lace1-deficient mice exhibit increased lactate uptake while concurrently impeding lactate efflux in iWAT during CL challenge. Lactate can be
converted to pyruvate to increase Ucp1 expression via lactate dehydrogenase (LDH) in the mitochondria of iWAT and BAT24. LDHB functions to convert lactate to pyruvate; in contrast, LDHA
converts pyruvate to lactate25. Hence, we proceeded to assess the mRNA expression of _Ldhb_ and _Ldha_ in iWAT following CL challenge. The results revealed elevated _Ldhb_ mRNA expression in
the Lace1 KO mice, whereas _Ldha_ mRNA expression remained unchanged compared with that in the CL-treated controls (Fig. 4f). Under cold conditions for 7 days, _Mct1_ mRNA levels, but not
_Mct4 levels_, in the iWAT of the Lace1 KO mice showed a trend toward increased expression, as in CL conditions (Supplementary Fig. 6a). In addition, _Ldhb_ but not Ldha mRNA expression was
increased (Supplementary Fig. 6b and c). Collectively, these findings suggest that Lace1-deficient mice exhibit increased lactate uptake in their iWAT following CL challenge or cold
exposure, with subsequent active conversion to pyruvate. For comprehensive verification, we performed Lace1 gene knockdown in a differentiated BAT cell line, followed by the induction of
adrenergic receptor activity through isoproterenol stimulation. Intriguingly, this manipulation resulted in elevated Mct1 gene expression, mirroring the in vivo observations (Fig. 4g and h).
Furthermore, when L-lactate was administered once daily for 3 days to 7-week-old male C57BL/6N mice, we noted a significant increase in _Ucp1_ protein and mRNA expression within the iWAT
(Fig. 4i–k). L-lactate, a well-recognized metabolite produced from pyruvate during anaerobic glycolysis26, has gained prominence as a novel metabolic regulator, with recent studies
highlighting its role in white adipose tissue browning23,24. Lace1 knockout in mice is thought to regulate lactate metabolism, potentially through the induction of white fat browning. LOSS
OF LACE1 RESULTS IN INACTIVATION OF PYRUVATE DEHYDROGENASE, WHICH PROMOTES LACTATE EFFLUX IN CL-INDUCED HEARTS To investigate why Lace1 KO mice have a high amount of circulating L-lactate
upon CL challenge, we measured _Mct4_ mRNA expression, which is related to a lactate efflux, in whole tissues (Fig. 5b). Lace1 KO mice presented highly increased _Mct4_ mRNA and protein
expression in the heart (Fig. 5b–d). However, MCT4 protein expression, which was significantly increased in the hearts of the CL-treated KO mice, did not differ in the soleus, gas, or kidney
(Supplementary Fig. 7). On the basis of the _Mct4_ mRNA level in the hearts of the Lace1 KO mice subjected to CL challenge, we hypothesized that lactate released from the heart contributes
to lactate uptake and utilization for browning in Lace1 KO mice subjected to CL challenge. Lactate release through Mct4 activation in heart tissue is a cardiac hypertrophic phenotype27.
Cardiac hypertrophy can progress to heart failure through increasing cardiomyocyte and heart size27,28. However, hypertrophic morphology was not observed in the hearts of the Lace1 KO mice
subjected to CL challenge. Nevertheless, we found that the hearts of the Lace1 KO mice displayed pyruvate dehydrogenase (PDH) inactivation compared with those of the WT mice under CL
challenge (Fig. 5e and f). When PDH is inactive, pyruvate from glycolysis cannot be converted to acetyl-CoA, thereby converting pyruvate to lactate. With an unbalanced pyruvate‒lactate axis,
lactate is released into the blood27 (Fig. 5a). Additionally, _Ldha_ mRNA levels were increased in the hearts of the Lace1 KO mice under CL challenge (Fig. 5g). This finding is in stark
contrast to the observed increase in MCT1 and LDHB levels following CL stimulation in iWAT. Taken together, our data revealed the inactivation of PDH in the hearts of the Lace1 KO mice
during CL challenge, alongside increased lactate efflux through MCT4 via increased LDHA expression. Moreover, we propose that these mechanisms may have contributed to lactate uptake-induced
browning in iWAT (Fig. 6). DISCUSSION The current study examined the role of Lace1 in the browning of iWAT. Lace1 expression was substantially increased in iWAT by browning stimuli such as
CL injection and cold exposure. However, the browning capacity of the Lace1-deficient mice was not disrupted; rather, the Lace1-deficient mice presented increased browning capacity. To
understand these conflicting consequences, we investigated the mechanisms by which Lace1 deficiency promotes browning capacity in mice. Here, we observed that CL-induced upregulation of
browning in Lace1-deficient mice was stimulated by lactate uptake in iWAT, suggesting for the first time that this phenomenon is due to increased lactate secretion induced by PDH
phosphorylation in the hearts of Lace1 KO mice. First, our data clearly revealed the expression of Lace1 in the browning process of iWAT, such as CL stimulation and cold exposure, which was
significantly correlated with the Ucp1 level. Ucp1 is the most representative marker of adipose tissue browning and is predominantly expressed in cells where many mitochondria are found,
such as BAT3,29. Thus, accumulating evidence has revealed the mechanism of Ucp1 expression in brown adipocytes and beige adipocytes with a high content of mitochondria and its effect on
whole-body energy homeostasis. Therefore, we concluded that Lace1 is a gene that plays an important role in the browning process. β3-ARs are the predominant regulators of WAT browning, and
CL is a potent and highly selective β3-AR agonist30. According to many studies related to WAT browning, CL treatment significantly increased browning marker gene expression, reduced lipid
droplet size, and increased heat generation and energy expenditure in both BAT and iWAT31,32, which was more evident over time. Since we identified Lace1 as an important gene for browning,
we administered CL to Lace1 KO mice and investigated the importance of Lace1 in the browning process. Interestingly, the Lace1 KO mice presented smaller lipid droplets and greater Ucp1
expression following CL treatment. Furthermore, oxygen uptake capacity and heat generation were significantly greater in the KO mice than in the WT mice. This phenomenon was the same with
cold exposure. To identify the cause or mediator of these unexpected results, we performed RNA-seq to investigate which gene had altered expression in CL-induced iWAT in Lace1 KO mice. We
found that the expression of the _Hcar1_ gene was the highest in iWAT when the CL was administered to Lace1 KO mice, which prompted us to establish a new hypothesis in this study. Hcar1,
also known as GPR81, is a cell-surface receptor for lactate and is highly expressed in adipose tissues but is also found in the kidney, skeletal muscle, and liver in mammals23.
Lactate-induced GPR81 effectively suppresses lipolysis via modulation of the cyclic adenosine monophosphate (cAMP)-protein kinase A (PKA) pathway, which contributes to hormone-sensitive
lipase (HSL) downregulation in adipose tissue32,33. Furthermore, the activation of the GPR81-induced cAMP‒PKA pathway is synergistically dependent on antilipolytic effects and insulin
actions34. These results suggest that lactate can modulate metabolic processes via Gpr81-dependent mechanisms in adipose tissue. Lactate may play a role in browning as a metabolite. Recent
studies reported that lactate plays a crucial role in the browning of WAT through its receptor GPR81 and transporter Mct1 in a β3-AR stimulation-dependent manner23,35. On the basis of a
previous study and our RNA-seq data, we hypothesized that CL-induced Hcar1 gene expression in the iWAT of the Lace1 KO mice could be due to increased circulating lactate levels. As expected,
the serum lactate level was significantly greater in the CL-induced Lace1 KO mice than in the WT mice, and the _Hcar1_ receptor level was significantly elevated in the Lace KO mice. In
addition, we confirmed the lactate uptake ability of the Lace1 KO mice by CL stimulation and confirmed that Mct1, a lactate influx transporter, was specifically expressed only in iWAT.
Lagarde et al. reported a positive correlation between Mct1 and thermogenic marker gene levels in iWAT. These researchers also revealed that Ucp1-positive cells were highly expressed in the
subpopulation of adipocytes expressing Mct1 after cold exposure. Lactate is first converted to pyruvate by the LDH reaction for oxidation, indicating that lactate consumption is directly
dependent on mitochondrial pyruvate utilization26,36. Compared with the WT mice, the CL-induced Lace1 KO mice presented significantly elevated Ldhb expression levels in iWAT. These results
suggest that CL-stimulated WAT from Lace1 KO mice takes up higher levels of lactate via upregulation of Mct1 than does that from WT mice and increases Ucp1 expression by increasing oxidative
phosphorylation in mitochondria through a high rate of conversion to pyruvate. We believe that these data provide essential insight into the role of lactate in β3-AR-induced browning of
iWAT. Although we found that Lace1 KO mice displayed increased browning capacity through lactate uptake upon CL stimulation, lactate concentrations did not differ in iWAT between WT and KO
mice. Therefore, we aimed to determine where and why lactate increases, and we measured Mct4, a lactate exporter, in whole tissues of the CL-stimulated WT mice and KO mice via the same
method as that used for Mct1 in iWAT. Interestingly, in contrast to the Mct1 results in iWAT, the CL-induced Lace1 KO mice exhibited exclusively increased Mct4 expression in the heart
compared with the WT mice. Under normal conditions, cardiomyocytes express low levels of Mct4, whose levels are increased by hypertrophy or heart failure27,28. However, we did not observe
hypertrophic morphology in the CL-induced Lace1 KO mice. Unlike previous reports, there was no difference in the cardiac hypertrophic phenotype according to elevated Mct4. Nevertheless, we
noted that PDH phosphorylation was significantly increased only in the Lace1 KO mice upon CL treatment. PDH is the rate-limiting enzyme for glucose oxidation, and cardiac-specific deletion
of PDH impairs glucose oxidation in mice37. Our results suggest that the increase in MCT4 in the CL-induced Lace1 KO mice is due to impaired pyruvate flux into mitochondria caused by PDH
phosphorylation. The role of β3-ARs in the heart has long been debated. This receptor could have either a protective effect or a detrimental effect on the heart38,39. However, studies on the
effect of CL-specific β3-AR activation on the heart have not been conducted, which is a limitation of the discussion in our study. Further studies are needed to investigate the molecular
mechanisms that may explain how LACE1 KO blocks PDH in a cardiac-specific manner and the mechanisms that increase MCT4 expression. These results would ultimately provide an important basis
for explaining the lactate-induced browning of iWAT observed in this study and could be the start of a new line of research to elucidate the relationship between cardiac metabolism and
adipocyte browning. Ikeda et al. suggested two reasons for the importance of beige fat studies. First, beige fat is a unique model for understanding how environmental factors control cell
fate and maintenance. Second, beige fat is relevant to adult humans and may be a therapeutic target in the treatment of metabolic disorders40. We first suggested that Lace1 is a new gene
that regulates the browning of white adipose tissue, which is the main target tissue of obesity metabolic research. Furthermore, we first suggested that CL, a specific agonist of β3-ARs,
which we used as a representative tool in browning research, regulates the browning of white fat by modulating the molecular and physiological functions of the heart through the Lace1 gene.
Therefore, the Lace1 gene is expected to play a role as an important therapeutic target in the future study of adipose tissue browning, obesity and metabolism. REFERENCES * Kaisanlahti, A.
& Glumoff, T. Browning of white fat: agents and implications for beige adipose tissue to type 2 diabetes. _J. Physiol. Biochem._ 75, 1–10 (2019). Article PubMed CAS Google Scholar *
Ishibashi, J. & Seale, P. Medicine. Beige can be slimming. _Science_ 328, 1113–1114 (2010). Article PubMed PubMed Central CAS Google Scholar * Kajimura, S. & Saito, M. A new era
in brown adipose tissue biology: molecular control of brown fat development and energy homeostasis. _Annu. Rev. Physiol._ 76, 225–249 (2014). Article PubMed CAS Google Scholar * Cohen,
P. & Spiegelman, B. M. Brown and beige fat: molecular parts of a thermogenic machine. _Diabetes_ 64, 2346–2351 (2015). Article PubMed PubMed Central CAS Google Scholar * Kajimura,
S., Spiegelman, B. M. & Seale, P. Brown and beige fat: physiological roles beyond heat generation. _Cell Metab._ 22, 546–559 (2015). Article PubMed PubMed Central CAS Google Scholar
* Hattori, K. et al. ASK1 signalling regulates brown and beige adipocyte function. _Nat. Commun._ 7, 11158 (2016). Article PubMed PubMed Central CAS Google Scholar * Shuai, L. et al.
SIRT5 regulates brown adipocyte differentiation and browning of subcutaneous white adipose tissue. _Diabetes_ 68, 1449–1461 (2019). Article PubMed CAS Google Scholar * Zhang, J. et al.
Dlgap1 negatively regulates browning of white fat cells through effects on cell proliferation and apoptosis. _Lipids Health Dis._ 19, 39 (2020). Article PubMed PubMed Central CAS Google
Scholar * Cesnekova, J. et al. The mammalian homologue of yeast Afg1 ATPase (lactation elevated 1) mediates degradation of nuclear-encoded complex IV subunits. _Biochem J._ 473, 797–804
(2016). Article PubMed CAS Google Scholar * Okuno, D., Iino, R. & Noji, H. Rotation and structure of FoF1-ATP synthase. _J. Biochem._ 149, 655–664 (2011). Article PubMed CAS
Google Scholar * Zheng, J. & Ramirez, V. D. Inhibition of mitochondrial proton F0F1-ATPase/ATP synthase by polyphenolic phytochemicals. _Br. J. Pharm._ 130, 1115–1123 (2000). Article
CAS Google Scholar * Li, Y. et al. MFSD7C switches mitochondrial ATP synthesis to thermogenesis in response to heme. _Nat. Commun._ 11, 4837 (2020). Article PubMed PubMed Central CAS
Google Scholar * Ikeda, K. et al. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. _Nat. Med._ 23,
1454–1465 (2017). Article PubMed PubMed Central CAS Google Scholar * Kim, Y. J. et al. Aerobic exercise for eight weeks provides protective effects towards liver and cardiometabolic
health and adipose tissue remodeling under metabolic stress for one week: a study in mice. _Metabolism_ 130, 155178 (2022). Article PubMed CAS Google Scholar * Aune U. L., Ruiz L. &
Kajimura S. Isolation and differentiation of stromal vascular cells to beige/brite cells. J. Vis. Exp. 50191 (2013). * Song, J., Pfanner, N. & Becker, T. Assembling the mitochondrial ATP
synthase. _Proc. Natl Acad. Sci. USA_ 115, 2850–2852 (2018). Article PubMed PubMed Central CAS Google Scholar * Razzoli, M., Emmett, M. J., Lazar, M. A. & Bartolomucci, A.
beta-Adrenergic receptors control brown adipose UCP-1 tone and cold response without affecting its circadian rhythmicity. _FASEB J._ 32, 5640–5646 (2018). Article PubMed PubMed Central
CAS Google Scholar * Hankir, M. K., Cowley, M. A. & Fenske, W. K. A BAT-centric approach to the treatment of diabetes: turn on the brain. _Cell Metab._ 24, 31–40 (2016). Article
PubMed CAS Google Scholar * Cypess, A. M. & Kahn, C. R. The role and importance of brown adipose tissue in energy homeostasis. _Curr. Opin. Pediatr._ 22, 478–484 (2010). Article
PubMed PubMed Central Google Scholar * Inokuma, K. et al. Indispensable role of mitochondrial UCP1 for antiobesity effect of beta3-adrenergic stimulation. _Am. J. Physiol. Endocrinol.
Metab._ 290, E1014–E1021 (2006). Article PubMed CAS Google Scholar * Lee, D. K. et al. Discovery and mapping of ten novel G protein-coupled receptor genes. _Gene_ 275, 83–91 (2001).
Article PubMed CAS Google Scholar * Roland, C. L. et al. Cell surface lactate receptor GPR81 is crucial for cancer cell survival. _Cancer Res_ 74, 5301–5310 (2014). Article PubMed
PubMed Central CAS Google Scholar * Yao, Z. et al. Dietary lactate supplementation protects against obesity by promoting adipose browning in mice. _J. Agric Food Chem._ 68, 14841–14849
(2020). Article PubMed CAS Google Scholar * Carriere, A. et al. Browning of white adipose cells by intermediate metabolites: an adaptive mechanism to alleviate redox pressure. _Diabetes_
63, 3253–3265 (2014). Article PubMed CAS Google Scholar * Urbanska K., Orzechowski A. Unappreciated role of LDHA and LDHB to control apoptosis and autophagy in tumor cells. Int J Mol
Sci. 20, 2085 (2019). * Brooks, G. A. The science and translation of lactate shuttle theory. _Cell Metab._ 27, 757–785 (2018). Article PubMed CAS Google Scholar * Cluntun, A. A. et al.
The pyruvate-lactate axis modulates cardiac hypertrophy and heart failure. _Cell Metab._ 33, 629–48 e10 (2021). Article PubMed CAS Google Scholar * Zhu, Y., Wu, J. & Yuan, S. Y. MCT1
and MCT4 expression during myocardial ischemic-reperfusion injury in the isolated rat heart. _Cell Physiol. Biochem._ 32, 663–674 (2013). Article PubMed CAS Google Scholar * Harms, M.
& Seale, P. Brown and beige fat: development, function and therapeutic potential. _Nat. Med._ 19, 1252–1263 (2013). Article PubMed CAS Google Scholar * Grujic, D. et al.
Beta3-adrenergic receptors on white and brown adipocytes mediate beta3-selective agonist-induced effects on energy expenditure, insulin secretion, and food intake. A study using transgenic
and gene knockout mice. _J. Biol. Chem._ 272, 17686–17693 (1997). Article PubMed CAS Google Scholar * Xiao, C., Goldgof, M., Gavrilova, O. & Reitman, M. L. Anti-obesity and metabolic
efficacy of the beta3-adrenergic agonist, CL316243, in mice at thermoneutrality compared to 22 degrees C. _Obesity_ 23, 1450–1459 (2015). Article PubMed CAS Google Scholar * Ge, H. et
al. Elucidation of signaling and functional activities of an orphan GPCR, GPR81. _J. Lipid Res._ 49, 797–803 (2008). Article PubMed CAS Google Scholar * Cai, T. Q. et al. Role of GPR81
in lactate-mediated reduction of adipose lipolysis. _Biochem. Biophys. Res. Commun._ 377, 987–991 (2008). Article PubMed CAS Google Scholar * Langin, D. Adipose tissue lipolysis
revisited (again!): lactate involvement in insulin antilipolytic action. _Cell Metab._ 11, 242–243 (2010). Article PubMed CAS Google Scholar * Lagarde, D. et al. Lactate fluxes mediated
by the monocarboxylate transporter-1 are key determinants of the metabolic activity of beige adipocytes. _J. Biol. Chem._ 296, 100137 (2021). Article PubMed CAS Google Scholar * Glancy,
B. et al. Mitochondrial lactate metabolism: history and implications for exercise and disease. _J. Physiol._ 599, 863–888 (2021). Article PubMed CAS Google Scholar * Gopal, K. et al.
Cardiac-specific deletion of pyruvate dehydrogenase impairs glucose oxidation rates and induces diastolic dysfunction. _Front. Cardiovasc. Med._ 5, 17 (2018). Article PubMed PubMed Central
Google Scholar * Moniotte, S. & Balligand, J. L. Potential use of beta(3)-adrenoceptor antagonists in heart failure therapy. _Cardiovasc. Drug Rev._ 20, 19–26 (2002). Article PubMed
CAS Google Scholar * Arioglu-Inan, E., Kayki-Mutlu, G. & Michel, M. C. Cardiac beta3 -adrenoceptors-A role in human pathophysiology? _Br. J. Pharm._ 176, 2482–2495 (2019). Article
CAS Google Scholar * Ikeda, K., Maretich, P. & Kajimura, S. The common and distinct features of brown and beige adipocytes. _Trends Endocrinol. Metab._ 29, 191–200 (2018). Article
PubMed PubMed Central CAS Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by the Korea Mouse Phenotyping Project (NRF-2013M3A9D5072550) of the Ministry of
Science and ICT through the National Research Foundation. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Laboratory of Developmental Biology and Genomics, Research Institute for Veterinary
Science, College of Veterinary Medicine, Seoul National University, Seoul, Republic of Korea Youn Ju Kim, Sang Gyu Lee, Hye Jin Kim & Je Kyung Seong * Korea Model animal Priority Center
(KMPC), Seoul National University, Seoul, Republic of Korea Youn Ju Kim, Sang Gyu Lee, Su In Jang, Hye Jin Kim & Je Kyung Seong * Division of Endocrine and Kidney Disease Research,
Department of Chronic Disease Convergence Research, Korea National Institute of Health, Korea Disease Control and Prevention Agency, Cheongju, Republic of Korea Youn Ju Kim *
Interdisciplinary Program in Bioinformatics, Seoul National University, Seoul, Republic of Korea Sang Gyu Lee & Je Kyung Seong * Metabolic Regulation Research Center, Korea Research
Institute of Bioscience and Biotechnology (KRIBB), Daejeon, Republic of Korea Won Kon Kim, Kyoung-Jin Oh & Kwang-Hee Bae * Department of Functional Genomics, KRIBB School of Bioscience,
Korea University of Science and Technology (UST), Daejeon, Republic of Korea Won Kon Kim, Kyoung-Jin Oh & Kwang-Hee Bae * Interdisciplinary Program for Bioinformatics, Program for Cancer
Biology, BIO-MAX/N-Bio Institute, Seoul National University, Seoul, Republic of Korea Je Kyung Seong Authors * Youn Ju Kim View author publications You can also search for this author
inPubMed Google Scholar * Sang Gyu Lee View author publications You can also search for this author inPubMed Google Scholar * Su In Jang View author publications You can also search for this
author inPubMed Google Scholar * Won Kon Kim View author publications You can also search for this author inPubMed Google Scholar * Kyoung-Jin Oh View author publications You can also
search for this author inPubMed Google Scholar * Kwang-Hee Bae View author publications You can also search for this author inPubMed Google Scholar * Hye Jin Kim View author publications You
can also search for this author inPubMed Google Scholar * Je Kyung Seong View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Youn Ju Kim:
Methodology, Data curation, Formal analysis, Investigation, Validation, Visualization, Writing—original draft. Sang Gyu Lee: Data curation, Formal analysis. Su In Jang: Methodology. Hye Jin
Kim: Data curation, Investigation, Validation, Visualization, Writing—review & editing. Je Kyung Seong: Funding acquisition, Project administration, Supervision. CORRESPONDING AUTHORS
Correspondence to Hye Jin Kim or Je Kyung Seong. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature
remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES RIGHTS AND PERMISSIONS OPEN ACCESS This
article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as
you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party
material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s
Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Kim, Y.J., Lee, S.G., Jang, S.I. _et al._
Lactate utilization in Lace1 knockout mice promotes browning of inguinal white adipose tissue. _Exp Mol Med_ 56, 2491–2502 (2024). https://doi.org/10.1038/s12276-024-01324-w Download
citation * Received: 04 January 2024 * Revised: 02 June 2024 * Accepted: 02 July 2024 * Published: 07 November 2024 * Issue Date: November 2024 * DOI:
https://doi.org/10.1038/s12276-024-01324-w SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative