Play all audios:
ABSTRACT In the present day, it is possible to incorporate targeted mutations or replace a gene using genome editing techniques such as customisable CRISPR/Cas9 system. Although induction of
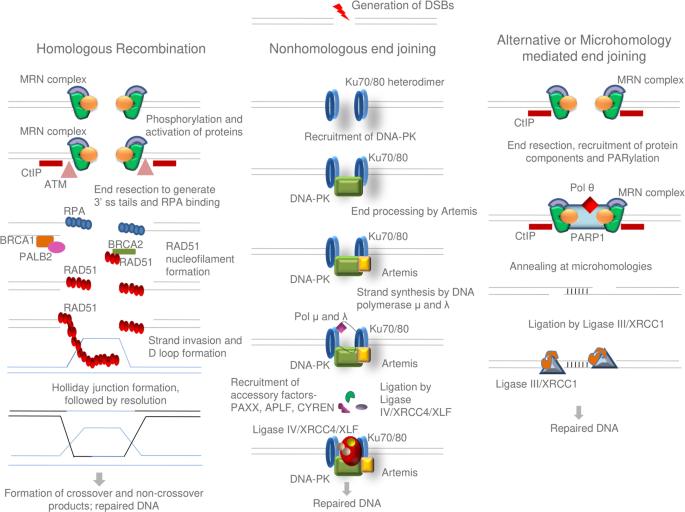
DNA double-strand breaks (DSBs) by genome editing tools can be repaired by both non-homologous end joining (NHEJ) and homologous recombination (HR), the skewness of the former pathway in
human and other mammals normally result in imprecise repair. Scientists working at the crossroads of DNA repair and genome editing have devised new strategies for using a specific pathway to
their advantage. Refinement in the efficiency of precise gene editing was witnessed upon downregulation of NHEJ by knockdown or using small molecule inhibitors on one hand, and upregulation
of HR proteins and addition of HR stimulators, other hand. The exploitation of cell cycle phase differences together with appropriate donor DNA length/sequence and small molecules has
provided further improvement in precise genome editing. The present article reviews the mechanisms of improving the efficiency of precise genome editing in several model organisms and in
clinics. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to
this journal Receive 50 print issues and online access $259.00 per year only $5.18 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy
now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer
support SIMILAR CONTENT BEING VIEWED BY OTHERS TARGETING DOUBLE-STRAND BREAK INDEL BYPRODUCTS WITH SECONDARY GUIDE RNAS IMPROVES CAS9 HDR-MEDIATED GENOME EDITING EFFICIENCIES Article Open
access 09 May 2022 SYNERGISTIC GENE EDITING IN HUMAN IPS CELLS VIA CELL CYCLE AND DNA REPAIR MODULATION Article Open access 08 June 2020 GENOME EDITING WITH THE HDR-ENHANCING DNA-PKCS
INHIBITOR AZD7648 CAUSES LARGE-SCALE GENOMIC ALTERATIONS Article Open access 27 November 2024 REFERENCES * Gaj T, Sirk SJ, Shui SL, Liu J. Genome-editing technologies: principles and
applications. Cold Spring Harb Perspect Biol. 2016;8:a023754. Article PubMed PubMed Central CAS Google Scholar * Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X. Applications of genome
editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther. 2020;5:1. Article PubMed PubMed Central Google Scholar *
Brinkman EK, Chen T, de Haas M, Holland HA, Akhtar W, van Steensel B. Kinetics and fidelity of the repair of Cas9-induced double-strand DNA breaks. Mol Cell. 2018;70:801–13.e806. Article
CAS PubMed PubMed Central Google Scholar * Jasin M, Haber JE. The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. DNA Repair.
2016;44:6–16. Article CAS PubMed PubMed Central Google Scholar * Mao Z, Bozzella M, Seluanov A, Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in
human cells. DNA Repair. 2008;7:1765–71. Article CAS PubMed PubMed Central Google Scholar * Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of
mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–8. Article CAS PubMed PubMed Central Google Scholar * Auer TO, Duroure K, De Cian
A, Concordet JP, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014;24:142–53. Article CAS PubMed PubMed Central
Google Scholar * Lackner DH, Carre A, Guzzardo PM, Banning C, Mangena R, Henley T, et al. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat Commun. 2015;6:10237. Article CAS
PubMed Google Scholar * Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–45. Article CAS PubMed Google Scholar * Javadekar SM, Raghavan SC.
Snaps and mends: DNA breaks and chromosomal translocations. FEBS J. 2015;282:2627–45. Article CAS PubMed Google Scholar * Khanna KK, Jackson SP. DNA double-strand breaks: signaling,
repair and the cancer connection. Nat Genet. 2001;27:247–54. Article CAS PubMed Google Scholar * Nambiar M, Raghavan SC. How does DNA break during chromosomal translocations? Nucleic
Acids Res. 2011;39:5813–25. Article CAS PubMed PubMed Central Google Scholar * Srivastava M, Raghavan SC. DNA double-strand break repair inhibitors as cancer therapeutics. Chem Biol.
2015;22:17–29. Article CAS PubMed Google Scholar * Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem.
2010;79:181–211. Article CAS PubMed PubMed Central Google Scholar * Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006;40:363–83. Article
CAS PubMed Google Scholar * Haber JE. A life investigating pathways that repair broken chromosomes. Annu Rev Genet. 2016;50:1–28. Article CAS PubMed Google Scholar * Heyer WD.
Regulation of recombination and genomic maintenance. Cold Spring Harb Perspect Biol. 2015;7:a016501. Article PubMed PubMed Central Google Scholar * Smith CE, Llorente B, Symington LS.
Template switching during break-induced replication. Nature. 2007;447:102–5. Article CAS PubMed Google Scholar * Bhargava R, Onyango DO, Stark JM. Regulation of single-strand annealing
and its role in genome maintenance. Trends Genet. 2016;32:566–75. Article CAS PubMed PubMed Central Google Scholar * Pandey MR. SC DNA double-strand break repair in mammals. J Radiat
Cancer Res. 2017;8:93–7. Article Google Scholar * Davis AJ, Chen DJ. DNA double strand break repair via non-homologous end-joining. Transl Cancer Res. 2013;2:130–43. CAS PubMed Google
Scholar * Grundy GJ, Rulten SL, Zeng Z, Arribas-Bosacoma R, Iles N, Manley K, et al. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J.
2013;32:112–25. Article CAS PubMed Google Scholar * Ochi T, Blackford AN, Coates J, Jhujh S, Mehmood S, Tamura N, et al. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku
to promote DNA double-strand break repair. Science. 2015;347:185–8. Article CAS PubMed PubMed Central Google Scholar * Hung PJ, Johnson B, Chen BR, Byrum AK, Bredemeyer AL, Yewdell WT,
et al. MRI is a DNA damage response adaptor during classical non-homologous end joining. Mol Cell. 2018;71:332–42.e338. Article CAS PubMed PubMed Central Google Scholar * Sharma S,
Javadekar SM, Pandey M, Srivastava M, Kumari R, Raghavan SC. Homology and enzymatic requirements of microhomology-dependent alternative end joining. Cell Death Dis. 2015;6:e1697. Article
CAS PubMed PubMed Central Google Scholar * Boboila C, Jankovic M, Yan CT, Wang JH, Wesemann DR, Zhang T, et al. Alternative end-joining catalyzes robust IgH locus deletions and
translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci USA. 2010;107:3034–9. Article CAS PubMed PubMed Central Google Scholar * Kent T, Chandramouly G, McDevitt
SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat Struct Mol Biol. 2015;22:230–7. Article CAS PubMed PubMed
Central Google Scholar * Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64. Article CAS PubMed
Google Scholar * Tomimatsu N, Mukherjee B, Catherine Hardebeck M, Ilcheva M, Vanessa Camacho C, Louise Harris J, et al. Phosphorylation of EXO1 by CDKs 1 and 2 regulates DNA end resection
and repair pathway choice. Nat Commun. 2014;5:3561. Article PubMed CAS Google Scholar * Wu Y, Kantake N, Sugiyama T, Kowalczykowski SC. Rad51 protein controls Rad52-mediated DNA
annealing. J Biol Chem. 2008;283:14883–92. Article CAS PubMed PubMed Central Google Scholar * Lok BH, Carley AC, Tchang B, Powell SN. RAD52 inactivation is synthetically lethal with
deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene. 2013;32:3552–8. Article CAS PubMed Google Scholar * Boch J, Scholze H,
Schornack S, Landgraf A, Hahn S, Kay S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–12. Article CAS PubMed Google Scholar *
Chandrasegaran S, Carroll D. Origins of programmable nucleases for genome engineering. J Mol Biol. 2016;428:963–89. Article CAS PubMed Google Scholar * Christian M, Cermak T, Doyle EL,
Schmidt C, Zhang F, Hummel A, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–61. Article CAS PubMed PubMed Central Google Scholar * Jinek
M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. Article CAS PubMed
PubMed Central Google Scholar * Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–60. Article
CAS PubMed PubMed Central Google Scholar * Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D. Requirements for double-strand cleavage by chimeric restriction
enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–9. Article CAS PubMed PubMed Central Google Scholar * Moscou MJ, Bogdanove AJ. A simple cipher governs
DNA recognition by TAL effectors. Science. 2009;326:1501. Article CAS PubMed Google Scholar * Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome
engineering via Cas9. Science. 2013;339:823–6. CAS PubMed PubMed Central Google Scholar * Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering.
Cell. 2014;157:1262–78. Article CAS PubMed PubMed Central Google Scholar * Savic N, Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Transl Res. 2016;168:15–21. Article
CAS PubMed Google Scholar * Moreno-Mateos MA, Fernandez JP, Rouet R, Vejnar CE, Lane MA, Mis E, et al. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled
genome editing. Nat Commun. 2017;8:2024. Article PubMed PubMed Central CAS Google Scholar * Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, et al. Efficient genome modification by
CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. Article CAS PubMed Google Scholar * Hyodo T, Rahman ML, Karnan S, Ito T, Toyoda A, Ota A, et al. Tandem
paired nicking promotes precise genome editing with scarce interference by p53. Cell Rep. 2020;30:1195–207.e1197. Article CAS PubMed Google Scholar * Ran FA, Hsu PD, Lin CY, Gootenberg
JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–9. Article CAS PubMed PubMed Central Google
Scholar * Beumer KJ, Trautman JK, Bozas A, Liu JL, Rutter J, Gall JG, et al. Efficient gene targeting in _Drosophila_ by direct embryo injection with zinc-finger nucleases. Proc Natl Acad
Sci USA. 2008;105:19821–6. Article CAS PubMed PubMed Central Google Scholar * Pawelczak KS, Gavande NS, VanderVere-Carozza PS, Turchi JJ. Modulating DNA repair pathways to improve
precision genome engineering. ACS Chem Biol. 2018;13:389–96. Article CAS PubMed Google Scholar * Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, et al.
Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett. 2004;14:6083–7. Article
CAS PubMed Google Scholar * Robert F, Barbeau M, Ethier S, Dostie J, Pelletier J. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med. 2015;7:93.
Article PubMed PubMed Central CAS Google Scholar * Dungl DA, Maginn EN, Stronach EA. Preventing damage limitation: targeting DNA-PKcs and DNA double-strand break repair pathways for
ovarian cancer therapy. Front Oncol. 2015;5:240. Article PubMed PubMed Central Google Scholar * Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, et al. In vivo genome
editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–9. Article CAS PubMed PubMed Central Google Scholar * Kostyushev D, Kostyusheva A,
Brezgin S, Zarifyan D, Utkina A, Goptar I, et al. Suppressing the NHEJ pathway by DNA-PKcs inhibitor NU7026 prevents degradation of HBV cccDNA cleaved by CRISPR/Cas9. Sci Rep. 2019;9:1847.
Article PubMed PubMed Central CAS Google Scholar * Riesenberg S, Chintalapati M, Macak D, Kanis P, Maricic T, Paabo S. Simultaneous precise editing of multiple genes in human cells.
Nucleic Acids Res. 2019;47:e116. Article CAS PubMed PubMed Central Google Scholar * Xu S, Kim J, Tang Q, Chen Q, Liu J, Xu Y, et al. CAS9 is a genome mutator by directly disrupting
DNA-PK dependent DNA repair pathway. Protein Cell. 2020;11:352–65. Article CAS PubMed PubMed Central Google Scholar * Saito S, Maeda R, Adachi N. Dual loss of human POLQ and LIG4
abolishes random integration. Nat Commun. 2017;8:16112. Article CAS PubMed PubMed Central Google Scholar * Zelensky AN, Schimmel J, Kool H, Kanaar R, Tijsterman M. Inactivation of Pol
theta and C-NHEJ eliminates off-target integration of exogenous DNA. Nat Commun. 2017;8:66. Article PubMed PubMed Central CAS Google Scholar * Boel A, De Saffel H, Steyaert W,
Callewaert B, De Paepe A, Coucke PJ, et al. CRISPR/Cas9-mediated homology-directed repair by ssODNs in zebrafish induces complex mutational patterns resulting from genomic integration of
repair-template fragments. Dis Model Mech. 2018;11:dmm 035352. Article CAS Google Scholar * Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, et al. Late embryonic lethality
and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–7. Article CAS PubMed Google Scholar * Srivastava M, Nambiar M, Sharma S, Karki SS, Goldsmith G,
Hegde M, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151:1474–87. Article CAS PubMed Google Scholar *
Kulashreshtha M, Mehta IS, Kumar P, Rao BJ. Chromosome territory relocation during DNA repair requires nuclear myosin 1 recruitment to chromatin mediated by Upsilon-H2AX signaling. Nucleic
Acids Res. 2016;44:8272–91. Article CAS PubMed PubMed Central Google Scholar * Reid DA, Keegan S, Leo-Macias A, Watanabe G, Strande NT, Chang HH, et al. Organization and dynamics of the
nonhomologous end-joining machinery during DNA double-strand break repair. Proc Natl Acad Sci USA. 2015;112:E2575–84. Article CAS PubMed PubMed Central Google Scholar * Reid DA, Conlin
MP, Yin Y, Chang HH, Watanabe G, Lieber MR, et al. Bridging of double-stranded breaks by the nonhomologous end-joining ligation complex is modulated by DNA end chemistry. Nucleic Acids Res.
2017;45:1872–8. Article CAS PubMed Google Scholar * Tripathi V, Agarwal H, Priya S, Batra H, Modi P, Pandey M, et al. MRN complex-dependent recruitment of ubiquitylated BLM helicase to
DSBs negatively regulates DNA repair pathways. Nat Commun. 2018;9:1016. Article PubMed PubMed Central CAS Google Scholar * Singh P, Schimenti JC, Bolcun-Filas E. A mouse geneticist’s
practical guide to CRISPR applications. Genetics. 2015;199:1–15. Article CAS PubMed Google Scholar * Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the
efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–42. Article CAS PubMed PubMed Central Google Scholar * Chu
VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat
Biotechnol. 2015;33:543–8. Article CAS PubMed Google Scholar * Ma Y, Chen W, Zhang X, Yu L, Dong W, Pan S, et al. Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing
in rats by inhibiting NHEJ and using Cas9 protein. RNA Biol. 2016;13:605–12. Article PubMed PubMed Central Google Scholar * Gutschner T, Haemmerle M, Genovese G, Draetta GF, Chin L.
Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Rep. 2016;14:1555–66. Article CAS PubMed Google Scholar * Yang D, Scavuzzo MA, Chmielowiec J,
Sharp R, Bajic A, Borowiak M. Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci Rep. 2016;6:21264.
Article CAS PubMed PubMed Central Google Scholar * Li G, Zhang X, Zhong C, Mo J, Quan R, Yang J, et al. Small molecules enhance CRISPR/Cas9-mediated homology-directed genome editing in
primary cells. Sci Rep. 2017;7:8943. Article PubMed PubMed Central CAS Google Scholar * Shao S, Ren C, Liu Z, Bai Y, Chen Z, Wei Z, et al. Enhancing CRISPR/Cas9-mediated
homology-directed repair in mammalian cells by expressing _Saccharomyces cerevisiae_ Rad52. Int J Biochem Cell Biol. 2017;92:43–52. Article CAS PubMed Google Scholar * Lin C, Li H, Hao
M, Xiong D, Luo Y, Huang C, et al. Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing of HSV-1 virus in human cells. Sci Rep. 2016;6:34531. Article CAS PubMed PubMed
Central Google Scholar * Hu Z, Shi Z, Guo X, Jiang B, Wang G, Luo D, et al. Ligase IV inhibitor SCR7 enhances gene editing directed by CRISPR-Cas9 and ssODN in human cancer cells. Cell
Biosci. 2018;8:12. Article PubMed PubMed Central CAS Google Scholar * Vartak SV, Swarup HA, Gopalakrishnan V, Gopinatha VK, Ropars V, Nambiar M, et al. Autocyclized and oxidized forms
of SCR7 induce cancer cell death by inhibiting nonhomologous DNA end joining in a Ligase IV dependent manner. FEBS J. 2018;285:3959–76. Article CAS PubMed Google Scholar * Greco GE,
Matsumoto Y, Brooks RC, Lu Z, Lieber MR, Tomkinson AE. SCR7 is neither a selective nor a potent inhibitor of human DNA ligase IV. DNA Repair. 2016;43:18–23. Article CAS PubMed PubMed
Central Google Scholar * Killian T, Dickopf S, Haas AK, Kirstenpfad C, Mayer K, Brinkmann U. Disruption of diphthamide synthesis genes and resulting toxin resistance as a robust technology
for quantifying and optimizing CRISPR/Cas9-mediated gene editing. Sci Rep. 2017;7:15480. Article PubMed PubMed Central CAS Google Scholar * Aslan Y, Tadjuidje E, Zorn AM, Cha SW.
High-efficiency non-mosaic CRISPR-mediated knock-in and indel mutation in F0 Xenopus. Development. 2017;144:2852–8. CAS PubMed PubMed Central Google Scholar * John F, George J,
Srivastava M, Hassan PA, Aswal VK, Karkie SS, et al. Pluronic copolymer encapsulated SCR7 as a potential anticancer agent. Faraday Discussions. 2015;177:155–61. Article CAS PubMed Google
Scholar * John F, George J, Vartak SV, Srivastava M, Hassan PA, Aswal VK, et al. Enhanced efficacy of pluronic copolymer micelle encapsulated SCR7 against cancer cell proliferation.
Macromol Biosci. 2015;15:521–34. Article CAS PubMed Google Scholar * Pandey M, Gopalakrishnan V, Swarup HA, Kumar S, Gudapureddy R, Jose AE, et al. Water-soluble version of SCR7-pyrazine
inhibits DNA repair and abrogates tumor cell proliferation. J Radiat Cancer Res. 2019;10:27–43. Article Google Scholar * Ray U, Jose AE, Suresh R, Kaloor U, Swarup HA, Nambiar M, et al.
Water-soluble SCR7 Can Abrogate DNA End Joining and Induce Cancer Cell Death. Clin Oncol Res. 2020;3:2–7. Google Scholar * Ray U, Raul SK, Gopinatha VK, Ghosh D, Rangappa KS, Mantelingu K,
et al. Identification and characterization of novel SCR7-based small-molecule inhibitor of DNA end-joining, SCR130 and its relevance in cancer therapeutics. Mol Carcinog. 2020;59:618–28.
Article CAS PubMed Google Scholar * Riesenberg S, Maricic T. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat Commun.
2018;9:2164. Article PubMed PubMed Central CAS Google Scholar * Pinder J, Salsman J, Dellaire G. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule
enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015;43:9379–92. Article CAS PubMed PubMed Central Google Scholar * Song J, Yang D, Xu J, Zhu T, Chen YE, Zhang J. RS-1
enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun. 2016;7:10548. Article CAS PubMed PubMed Central Google Scholar * Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, et
al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–7. Article CAS PubMed PubMed Central Google Scholar * Davis KM, Pattanayak V,
Thompson DB, Zuris JA, Liu DR. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol. 2015;11:316–8. Article CAS PubMed PubMed Central Google
Scholar * Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766. Article PubMed
PubMed Central Google Scholar * Plaza Reyes A, Lanner F. Time matters: gene editing at the mouse 2-cell embryo stage boosts knockin efficiency. Cell Stem Cell. 2018;23:155–7. Article CAS
PubMed Google Scholar * Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature.
2015;528:422–6. Article CAS PubMed PubMed Central Google Scholar * Charpentier M, Khedher AHY, Menoret S, Brion A, Lamribet K, Dardillac E, et al. CtIP fusion to Cas9 enhances transgene
integration by homology-dependent repair. Nat Commun. 2018;9:1133. Article CAS PubMed PubMed Central Google Scholar * Tran NT, Bashir S, Li X, Rossius J, Chu VT, Rajewsky K, et al.
Enhancement of precise gene editing by the association of Cas9 with homologous recombination factors. Front Genet. 2019;10:365. Article CAS PubMed PubMed Central Google Scholar *
Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–46. Article CAS
PubMed PubMed Central Google Scholar * Song F, Stieger K. Optimizing the DNA donor template for homology-directed repair of double-strand breaks. Mol Ther Nucleic Acids. 2017;7:53–60.
Article CAS PubMed PubMed Central Google Scholar * Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by
CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–9. Article CAS PubMed PubMed Central Google Scholar * Mikuni T, Nishiyama J, Sun Y, Kamasawa N, Yasuda R. High-throughput,
high-resolution mapping of protein localization in mammalian brain by in vivo genome editing. Cell. 2016;165:1803–17. Article CAS PubMed PubMed Central Google Scholar * Miura H, Quadros
RM, Gurumurthy CB, Ohtsuka M. Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat Protoc. 2018;13:195–215. Article CAS PubMed Google
Scholar * Aird EJ, Lovendahl KN, St Martin A, Harris RS, Gordon WR. Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun
Biol. 2018;1:54. * Carlson-Stevermer J, Abdeen AA, Kohlenberg L, Goedland M, Molugu K, Lou M, et al. Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA
aptamer for precise gene editing. Nat Commun. 2017;8:1711. Article PubMed PubMed Central CAS Google Scholar * Ma M, Zhuang F, Hu X, Wang B, Wen XZ, Ji JF, et al. Efficient generation of
mice carrying homozygous double-floxp alleles using the Cas9-Avidin/Biotin-donor DNA system. Cell Res. 2017;27:578–81. Article CAS PubMed PubMed Central Google Scholar * Savic N,
Ringnalda FC, Lindsay H, Berk C, Bargsten K, Li Y, et al. Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. Elife 2018;7. * Chen S,
Sun S, Moonen D, Lee C, Lee AY, Schaffer DV, et al. CRISPR-READI: efficient generation of knockin mice by CRISPR RNP electroporation and AAV donor infection. Cell Rep. 2019;27:3780–89.e3784.
Article CAS PubMed PubMed Central Google Scholar * Paulsen BS, Mandal PK, Frock RL, Boyraz B, Yadav R, Upadhyayula S, et al. Ectopic expression of RAD52 and dn53BP1 improves
homology-directed repair during CRISPR-Cas9 genome editing. Nat Biomed Eng. 2017;1:878–88. Article CAS PubMed PubMed Central Google Scholar * Ochs F, Somyajit K, Altmeyer M, Rask MB,
Lukas J, Lukas C. 53BP1 fosters fidelity of homology-directed DNA repair. Nat Struct Mol Biol. 2016;23:714–21. Article CAS PubMed Google Scholar * Canny MD, Moatti N, Wan LCK,
Fradet-Turcotte A, Krasner D, Mateos-Gomez PA, et al. Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency. Nat Biotechnol.
2018;36:95–102. Article CAS PubMed Google Scholar * Jayavaradhan R, Pillis DM, Goodman M, Zhang F, Zhang Y, Andreassen PR, et al. CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances
HDR and inhibits NHEJ specifically at Cas9 target sites. Nat Commun. 2019;10:2866. Article PubMed PubMed Central CAS Google Scholar * Di Primio C, Galli A, Cervelli T, Zoppe M, Rainaldi
G. Potentiation of gene targeting in human cells by expression of _Saccharomyces cerevisiae_ Rad52. Nucleic Acids Res. 2005;33:4639–48. Article PubMed PubMed Central CAS Google Scholar
* Davis L, Maizels N. Two distinct pathways support gene correction by single-stranded donors at DNA Nicks. Cell Rep. 2016;17:1872–81. Article CAS PubMed PubMed Central Google Scholar
* Richardson CD, Kazane KR, Feng SJ, Zelin E, Bray NL, Schafer AJ, et al. CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat Genet. 2018;50:1132–9.
Article CAS PubMed Google Scholar * Nambiar TS, Billon P, Diedenhofen G, Hayward SB, Taglialatela A, Cai K, et al. Stimulation of CRISPR-mediated homology-directed repair by an
engineered RAD18 variant. Nat Commun. 2019;10:3395. Article PubMed PubMed Central CAS Google Scholar * Yu S, Song Z, Luo J, Dai Y, Li N. Over-expression of RAD51 or RAD54 but not
RAD51/4 enhances extra-chromosomal homologous recombination in the human sarcoma (HT-1080) cell line. J Biotechnol. 2011;154:21–4. Article CAS PubMed Google Scholar * Ye L, Wang C, Hong
L, Sun N, Chen D, Chen S, et al. Programmable DNA repair with CRISPRa/i enhanced homology-directed repair efficiency with a single Cas9. Cell Discov. 2018;4:46. Article PubMed PubMed
Central CAS Google Scholar * Wienert B, Nguyen DN, Guenther A, Feng SJ, Locke MN, Wyman SK, et al. Timed inhibition of CDC7 increases CRISPR-Cas9 mediated templated repair. Nat Commun.
2020;11:2109. Article CAS PubMed PubMed Central Google Scholar * Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, et al. Human hematopoietic stem/progenitor cells modified by
zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–47. Article CAS PubMed PubMed Central Google Scholar * Barrangou R, Doudna JA. Applications of
CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34:933–41. Article CAS PubMed Google Scholar * Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, et
al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–26. Article CAS PubMed Google Scholar * Allen AG, Chung CH, Atkins
A, Dampier W, Khalili K, Nonnemacher MR, et al. Gene editing of HIV-1 co-receptors to prevent and/or cure virus infection. Front Microbiol. 2018;9:2940. Article PubMed PubMed Central
Google Scholar * Song B, Fan Y, He W, Zhu D, Niu X, Wang D, et al. Improved hematopoietic differentiation efficiency of gene-corrected beta-thalassemia induced pluripotent stem cells by
CRISPR/Cas9 system. Stem Cells Dev. 2015;24:1053–65. Article CAS PubMed Google Scholar * Huang X, Wang Y, Yan W, Smith C, Ye Z, Wang J, et al. Production of gene-corrected adult beta
globin protein in human erythrocytes differentiated from patient iPSCs after genome editing of the sickle point mutation. Stem Cells. 2015;33:1470–9. Article CAS PubMed PubMed Central
Google Scholar * Smith C, Abalde-Atristain L, He C, Brodsky BR, Braunstein EM, Chaudhari P, et al. Efficient and allele-specific genome editing of disease loci in human iPSCs. Mol Ther.
2015;23:570–7. Article CAS PubMed Google Scholar * Shim G, Kim D, Park GT, Jin H, Suh SK, Oh YK. Therapeutic gene editing: delivery and regulatory perspectives. Acta Pharmacol Sin.
2017;38:738–53. Article CAS PubMed PubMed Central Google Scholar * Du J, Shang J, Chen F, Zhang Y, Yin N, Xie T, et al. A CRISPR/Cas9-based screening for non-homologous end joining
inhibitors reveals ouabain and penfluridol as radiosensitizers. Mol Cancer Ther. 2018;17:419–31. Article CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We thank Urbi Roy,
Dipayan Ghosh and other members of SCR laboratory for critical reading and comments on the paper. This work was supported by grants from CEFIPRA (IFC/5203-4/2015/131), DAE
(21/01/2016-BRNS/35074), DBT Glue-Grant (BT/PR23078/MED/29/1253/2017), IISc-DBT partnership programme (BT/PR27952-INF/22/212/2018) to SCR. UR is supported by Senior Research Fellowship (SRF)
from CSIR, India. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Biochemistry, Indian Institute of Science, Bangalore, 560012, India Ujjayinee Ray & Sathees C. Raghavan
Authors * Ujjayinee Ray View author publications You can also search for this author inPubMed Google Scholar * Sathees C. Raghavan View author publications You can also search for this
author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Sathees C. Raghavan. ETHICS DECLARATIONS CONFLICT OF INTEREST The authors declare that they have no conflict of
interest. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. RIGHTS AND
PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Ray, U., Raghavan, S.C. Modulation of DNA double-strand break repair as a strategy to improve precise genome
editing. _Oncogene_ 39, 6393–6405 (2020). https://doi.org/10.1038/s41388-020-01445-2 Download citation * Received: 06 May 2020 * Revised: 07 August 2020 * Accepted: 21 August 2020 *
Published: 03 September 2020 * Issue Date: 08 October 2020 * DOI: https://doi.org/10.1038/s41388-020-01445-2 SHARE THIS ARTICLE Anyone you share the following link with will be able to read
this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative