Play all audios:
ABSTRACT Graft-versus-host disease (GVHD) and cytomegalovirus (CMV)-related complications are leading causes of mortality after unrelated-donor hematopoietic cell transplantation (UD-HCT).
The non-conventional MHC class I gene _MICB_, alike _MICA_, encodes a stress-induced polymorphic NKG2D ligand. However, unlike MICA, MICB interacts with the CMV-encoded UL16, which
sequestrates MICB intracellularly, leading to immune evasion. Here, we retrospectively analyzed the impact of mismatches in MICB amino acid position 98 (MICB98), a key polymorphic residue
involved in UL16 binding, in 943 UD-HCT pairs who were allele-matched at _HLA-A_, _-B_, _-C_, _-DRB1_, _-DQB1_ and _MICA_ loci. _HLA-DP_ typing was further available. _MICB98_ mismatches
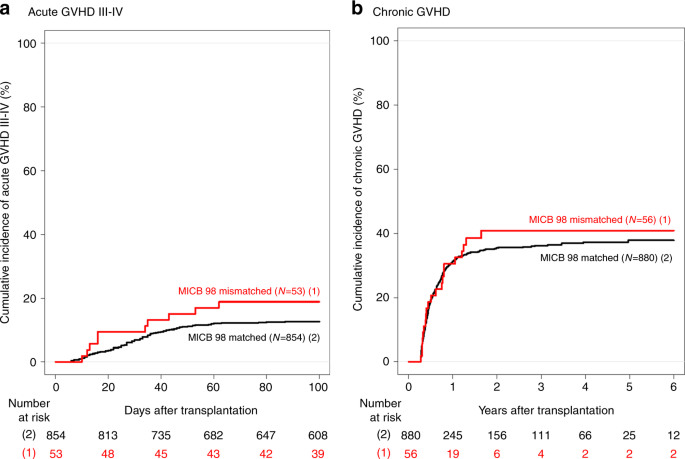
were significantly associated with an increased incidence of acute (grade II–IV: HR, 1.20; 95% CI, 1.15 to 1.24; _P_ < 0.001; grade III–IV: HR, 2.28; 95% CI, 1.56 to 3.34; _P_ < 0.001)
and chronic GVHD (HR, 1.21; 95% CI, 1.10 to 1.33; _P_ < 0.001). _MICB98_ matching significantly reduced the effect of CMV status on overall mortality from a hazard ratio of 1.77 to 1.16.
_MICB98_ mismatches showed a GVHD-independent association with a higher incidence of CMV infection/reactivation (HR, 1.84; 95% CI, 1.34 to 2.51; _P_ < 0.001). Hence selecting a
_MICB98_-matched donor significantly reduces the GVHD incidence and lowers the impact of CMV status on overall survival. Access through your institution Buy or subscribe This is a preview of
subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $259.00 per year only
$21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout
ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS IMPACT OF PATIENT: DONOR HLA
DISPARITY ON REDUCED-INTENSITY-CONDITIONED ALLOGENEIC STEM CELL TRANSPLANTS FROM HLA MISMATCHED UNRELATED DONORS FOR AML: FROM THE ALWP OF THE EBMT Article 02 October 2020 THE OUTCOME OF TWO
OR MORE HLA LOCI MISMATCHED UNRELATED DONOR HEMATOPOIETIC CELL TRANSPLANTATION FOR ACUTE LEUKEMIA: AN ALWP OF THE EBMT STUDY Article 19 June 2020 INDIVIDUAL HLAS INFLUENCE IMMUNOLOGICAL
EVENTS IN ALLOGENEIC STEM CELL TRANSPLANTATION FROM HLA-IDENTICAL SIBLING DONORS Article 09 October 2020 REFERENCES * Forman JS, Negrin SR, Antin HJ, Appelbaum RF. Thomas’ hematopoietic cell
transplantation, 5th ed.; Chichester: Wiley-blackwell; 2016. * Appelbaum FR, Thomas ED. Thomas' hematopoietic cell transplantation: stem cell transplantation, 4th ed. Wiley-Blackwell:
Chichester; 2009. * Gratwohl A, Baldomero H, Aljurf M, Pasquini MC, Bouzas LF, Yoshimi A, et al. Hematopoietic stem cell transplantation: a global perspective. JAMA. 2010;303:1617–24.
https://doi.org/10.1001/jama.2010.491. Article CAS PubMed PubMed Central Google Scholar * D'Souza A, Fretham C. Current uses and outcomes of hematopoietic cell transplantation
(HCT): CIBMTR Summary Slides; 2018. https://www.cibmtr.org. Accessed April 2019. * Gooley TA, Chien JW, Pergam SA, Hingorani S, Sorror ML, Boeckh M, et al. Reduced mortality after allogeneic
hematopoietic-cell transplantation. N Engl J Med. 2010;363:2091–101. https://doi.org/10.1056/NEJMoa1004383. Article CAS PubMed PubMed Central Google Scholar * Warren EH, Zhang XC, Li
S, Fan W, Storer BE, Chien JW, et al. Effect of MHC and non-MHC donor/recipient genetic disparity on the outcome of allogeneic HCT. Blood. 2012;120:2796–806.
https://doi.org/10.1182/blood-2012-04-347286. Article CAS PubMed PubMed Central Google Scholar * Mori T, Kato J. Cytomegalovirus infection/disease after hematopoietic stem cell
transplantation. Int J Hematol. 2010;91:588–95. https://doi.org/10.1007/s12185-010-0569-x. e-pub ahead of print 2010/04/24. Article PubMed Google Scholar * Paris C, Kopp K, King A,
Santolaya ME, Zepeda AJ, Palma J. Cytomegalovirus infection in children undergoing hematopoietic stem cell transplantation in Chile. Pediatr Blood Cancer. 2009;53:453–8.
https://doi.org/10.1002/pbc.22060. Article PubMed Google Scholar * Boeckh M, Nichols WG, Papanicolaou G, Rubin R, Wingard JR, Zaia J. Cytomegalovirus in hematopoietic stem cell transplant
recipients: current status, known challenges, and future strategies. Biol Blood Marrow Transplant. 2003;9:543–58. Article PubMed Google Scholar * Boeckh M, Ljungman P. How we treat
cytomegalovirus in hematopoietic cell transplant recipients. Blood. 2009;113:5711–9. https://doi.org/10.1182/blood-2008-10-143560. e-pub ahead of print 2009/03/21. Article CAS PubMed
PubMed Central Google Scholar * Ljungman P, Griffiths P, Paya C. Definitions of cytomegalovirus infection and disease in transplant recipients. Clin Infect Dis. 2002;34:1094–7.
https://doi.org/10.1086/339329. Article PubMed Google Scholar * Ljungman P, Hakki M, Boeckh M. Cytomegalovirus in hematopoietic stem cell transplant recipients. Hematol/Oncol Clin N Am.
2011;25:151–69. https://doi.org/10.1016/j.hoc.2010.11.011. Article Google Scholar * Takenaka K, Nishida T, Asano-Mori Y, Oshima K, Ohashi K, Mori T, et al. Cytomegalovirus reactivation
after allogeneic hematopoietic stem cell transplantation is associated with a reduced risk of relapse in patients with acute myeloid leukemia who survived to day 100 after transplantation:
the Japan Society for Hematopoietic Cell Transplantation Transplantation-related Complication Working Group. Biol Blood Marrow Transplant. 2015;21:2008–16.
https://doi.org/10.1016/j.bbmt.2015.07.019. Article PubMed Google Scholar * Manjappa S, Bhamidipati PK, Stokerl-Goldstein KE, DiPersio JF, Uy GL, Westervelt P, et al. Protective effect of
cytomegalovirus reactivation on relapse after allogeneic hematopoietic cell transplantation in acute myeloid leukemia patients is influenced by conditioning regimen. Biol Blood Marrow
Transplant. 2014;20:46–52. https://doi.org/10.1016/j.bbmt.2013.10.003. e-pub ahead of print 2013/10/15. Article CAS PubMed Google Scholar * Jang JE, Kim SJ, Cheong JW, Hyun SY, Kim YD,
Kim YR, et al. Early CMV replication and subsequent chronic GVHD have a significant anti-leukemic effect after allogeneic HSCT in acute myeloid leukemia. Ann Hematol. 2015;94:275–82.
https://doi.org/10.1007/s00277-014-2190-1. e-pub ahead of print 2014/08/20. Article CAS PubMed Google Scholar * Nichols WG, Corey L, Gooley T, Davis C, Boeckh M. High risk of death due
to bacterial and fungal infection among cytomegalovirus (CMV)-seronegative recipients of stem cell transplants from seropositive donors: evidence for indirect effects of primary CMV
infection. J Infect Dis. 2002;185:273–82. https://doi.org/10.1086/338624. e-pub ahead of print 2002/01/25. Article PubMed Google Scholar * Sousa H, Boutolleau D, Ribeiro J, Teixeira AL,
Pinho Vaz C, Campilho F, et al. Cytomegalovirus infection in patients who underwent allogeneic hematopoietic stem cell transplantation in Portugal: a five-year retrospective review. Biol
Blood Marrow Transplant. 2014;20:1958–67. https://doi.org/10.1016/j.bbmt.2014.08.010. Article PubMed Google Scholar * Ichihara H, Nakamae H, Hirose A, Nakane T, Koh H, Hayashi Y, et al.
Immunoglobulin prophylaxis against cytomegalovirus infection in patients at high risk of infection following allogeneic hematopoietic cell transplantation. Transplant Proc. 2011;43:3927–32.
https://doi.org/10.1016/j.transproceed.2011.08.104. Article CAS PubMed Google Scholar * Schmidt-Hieber M, Labopin M, Beelen D, Volin L, Ehninger G, Finke J, et al. CMV serostatus still
has an important prognostic impact in de novo acute leukemia patients after allogeneic stem cell transplantation: a report from the Acute Leukemia Working Party of EBMT. Blood.
2013;122:3359–64. https://doi.org/10.1182/blood-2013-05-499830. Article CAS PubMed Google Scholar * Petersdorf EW. Optimal HLA matching in hematopoietic cell transplantation. Curr Opin
Immunol. 2008;20:588–93. https://doi.org/10.1016/j.coi.2008.06.014. Article CAS PubMed PubMed Central Google Scholar * Miller W, Flynn P, McCullough J, Balfour HH,Jr, Goldman A, Haake
R, et al. Cytomegalovirus infection after bone marrow transplantation: an association with acute graft-v-host disease. Blood. 1986;67:1162–7. Article CAS PubMed Google Scholar *
Flomenberg N, Baxter-Lowe LA, Confer D, Fernandez-Vina M, Filipovich A, Horowitz M, et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone
marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood. 2004;104:1923–30. https://doi.org/10.1182/blood-2004-03-0803. Article
CAS PubMed Google Scholar * Ruell J, Barnes C, Mutton K, Foulkes B, Chang J, Cavet J, et al. Active CMV disease does not always correlate with viral load detection. Bone Marrow
Transplant. 2007;40:55–61. https://doi.org/10.1038/sj.bmt.1705671. Article CAS PubMed Google Scholar * Castagnola E, Cappelli B, Erba D, Rabagliati A, Lanino E, Dini G. Cytomegalovirus
infection after bone marrow transplantation in children. Hum Immunol. 2004;65:416–22. https://doi.org/10.1016/j.humimm.2004.02.013. Article PubMed Google Scholar * Bahram S, Bresnahan M,
Geraghty DE, Spies T. A second lineage of mammalian major histocompatibility complex class I genes. Proc Natl Acad Sci USA. 1994;91:6259–63. https://doi.org/10.1073/pnas.91.14.6259. Article
CAS PubMed PubMed Central Google Scholar * Carapito R, Bahram S. Genetics, genomics, and evolutionary biology of NKG2D ligands. Immunological Rev. 2015;267:88–116.
https://doi.org/10.1111/imr.12328. Article CAS Google Scholar * Bahram S, Spies T. Nucleotide sequence of a human MHC class I MICB cDNA. Immunogenetics. 1996;43:230–3. CAS PubMed Google
Scholar * Bahram S, Shiina T, Oka A, Tamiya G, Inoko H. Genomic structure of the human MHC class I MICB gene. Immunogenetics. 1996;45:161–2. Article CAS PubMed Google Scholar *
Carapito R, Jung N, Kwemou M, Untrau M, Michel S, Pichot A, et al. Matching for the nonconventional MHC-I MICA gene significantly reduces the incidence of acute and chronic GVHD. Blood.
2016;128:1979–86. https://doi.org/10.1182/blood-2016-05-719070. Article CAS PubMed PubMed Central Google Scholar * Fuerst D, Neuchel C, Niederwieser D, Bunjes D, Gramatzki M, Wagner E,
et al. Matching for the MICA-129 polymorphism is beneficial in unrelated hematopoietic stem cell transplantation. Blood. 2016;128:3169–76. https://doi.org/10.1182/blood-2016-05-716357.
Article CAS PubMed Google Scholar * Petersdorf EW, Hansen JA, Martin PJ, Woolfrey A, Malkki M, Gooley T, et al. Major-histocompatibility-complex class I alleles and antigens in
hematopoietic-cell transplantation. N Engl J Med. 2001;345:1794–800. https://doi.org/10.1056/NEJMoa011826. Article CAS PubMed Google Scholar * Groh V, Bahram S, Bauer S, Herman A,
Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci USA. 1996;93:12445–50. Article
CAS PubMed PubMed Central Google Scholar * Wang WY, Tian W, Zhu FM, Liu XX, Li LX, Wang F. MICA, MICB polymorphisms and linkage disequilibrium with HLA-B in a Chinese Mongolian
population. Scand J Immunol. 2016;83:456–62. https://doi.org/10.1111/sji.12437. Article CAS PubMed Google Scholar * Liu X, Tian W, Li L, Cai J. Characterization of the major
histocompatibility complex class I chain-related gene B (MICB) polymorphism in a northern Chinese Han population: the identification of a new MICB allele, MICB*023. Hum Immunol.
2011;72:727–32. https://doi.org/10.1016/j.humimm.2011.05.013. Article CAS PubMed Google Scholar * Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat
Immunol. 2008;9:495–502. https://doi.org/10.1038/ni1581. Article CAS PubMed PubMed Central Google Scholar * Cosman D, Mullberg J, Sutherland CL, Chin W, Armitage R, Fanslow W, et al.
ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–33. Article CAS PubMed Google
Scholar * Klumkrathok K, Jumnainsong A, Leelayuwat C. Allelic MHC class I chain related B (MICB) molecules affect the binding to the human cytomegalovirus (HCMV) unique long 16 (UL16)
protein: implications for immune surveillance. J Microbiol. 2013;51:241–6. https://doi.org/10.1007/s12275-013-2514-1. Article CAS PubMed Google Scholar * Muller S, Zocher G, Steinle A,
Stehle T. Structure of the HCMV UL16-MICB complex elucidates select binding of a viral immunoevasin to diverse NKG2D ligands. PLoS Pathog. 2010;6:e1000723.
https://doi.org/10.1371/journal.ppat.1000723. Article CAS PubMed PubMed Central Google Scholar * Hill GR, Ferrara JL. The primacy of the gastrointestinal tract as a target organ of
acute graft-versus-host disease: rationale for the use of cytokine shields in allogeneic bone marrow transplantation. Blood. 2000;95:2754–9. Article CAS PubMed Google Scholar *
Isernhagen A, Malzahn D, Viktorova E, Elsner L, Monecke S, von Bonin F, et al. The MICA-129 dimorphism affects NKG2D signaling and outcome of hematopoietic stem cell transplantation. EMBO
Mol Med. 2015;7:1480–502. https://doi.org/10.15252/emmm.201505246. Article CAS PubMed PubMed Central Google Scholar * Boukouaci W, Busson M, Peffault de Latour R, Rocha V, Suberbielle
C, Bengoufa D, et al. MICA-129 genotype, soluble MICA, and anti-MICA antibodies as biomarkers of chronic graft-versus-host disease. Blood. 2009;114:5216–24.
https://doi.org/10.1182/blood-2009-04-217430. Article CAS PubMed Google Scholar * Parmar S, Del Lima M, Zou Y, Patah PA, Liu P, Cano P, et al. Donor-recipient mismatches in MHC class I
chain-related gene A in unrelated donor transplantation lead to increased incidence of acute graft-versus-host disease. Blood. 2009;114:2884–7. https://doi.org/10.1182/blood-2009-05-223172.
Article CAS PubMed PubMed Central Google Scholar * Pellet P, Renaud M, Fodil N, Laloux L, Inoko H, Hauptmann G, et al. Allelic repertoire of the human MICB gene. Immunogenetics.
1997;46:434–6. Article CAS PubMed Google Scholar * Glucksberg H, Storb R, Fefer A, Buckner CD, Neiman PE, Clift RA, et al. Clinical manifestations of graft-versus-host disease in human
recipients of marrow from HL-A-matched sibling donors. Transplantation. 1974;18:295–304. https://doi.org/10.1097/00007890-197410000-00001. Article CAS PubMed Google Scholar * Scheike TH,
Zhang MJ. Analyzing competing risk data using the R timereg package. J Stat Softw. 2011;30:i02. Google Scholar * Scheike TH, Zhang MJ. Flexible competing risks regression modeling and
goodness-of-fit. Lifetime Data Anal. 2008;14:464–83. https://doi.org/10.1007/s10985-008-9094-0. Article PubMed PubMed Central Google Scholar * Scheike T, Zhang M, Gerds T. Predicting
cumulative incidence probability by direct binomial regression. Biometrika. 2008;95:205–20. Article Google Scholar * Therneau T, Grambsch P. Modeling survival data: extending the Cox
model. New York: Springer; 2000. * Fleischhauer K, Shaw BE, Gooley T, Malkki M, Bardy P, Bignon JD, et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor
haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13:366–74. https://doi.org/10.1016/S1470-2045(12)70004-9. Article CAS PubMed Google Scholar * Peduzzi P,
Concato J, Feinstein AR, Holford TR. Importance of events per independent variable in proportional hazards regression analysis. II. Accuracy and precision of regression estimates. J Clin
Epidemiol. 1995;48:1503–10. https://doi.org/10.1016/0895-4356(95)00048-8. Article CAS PubMed Google Scholar * Peduzzi P, Concato J, Kemper E, Holford TR, Feinstein AR. A simulation study
of the number of events per variable in logistic regression analysis. J Clin Epidemiol. 1996;49:1373–9. https://doi.org/10.1016/s0895-4356(96)00236-3. Article CAS PubMed Google Scholar
* Team RDC. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2010. http://R-project.org. * Wu J, Chalupny NJ, Manley TJ, Riddell SR,
Cosman D, Spies T. Intracellular retention of the MHC class I-related chain B ligand of NKG2D by the human cytomegalovirus UL16 glycoprotein. J Immunol. 2003;170:4196–200. Article CAS
PubMed Google Scholar * Spreu J, Stehle T, Steinle A. Human cytomegalovirus-encoded UL16 discriminates MIC molecules by their alpha2 domains. J Immunol. 2006;177:3143–9. Article CAS
PubMed Google Scholar * Nikolich-Zugich J, Goodrum F, Knox K, Smithey MJ. Known unknowns: how might the persistent herpesvirome shape immunity and aging? Curr Opin Immunol. 2017;48:23–30.
https://doi.org/10.1016/j.coi.2017.07.011. Article CAS PubMed PubMed Central Google Scholar * de la Camara R. CMV in hematopoietic stem cell transplantation. Mediterranean J Hematol
Infect Dis. 2016;8:e2016031. https://doi.org/10.4084/MJHID.2016.031. Article Google Scholar * Broers AE, van Der Holt R, van Esser JW, Gratama JW, Henzen-Logmans S, Kuenen-Boumeester V, et
al. Increased transplant-related morbidity and mortality in CMV-seropositive patients despite highly effective prevention of CMV disease after allogeneic T-cell-depleted stem cell
transplantation. Blood. 2000;95:2240–5. Article CAS PubMed Google Scholar * Cantoni N, Hirsch HH, Khanna N, Gerull S, Buser A, Bucher C, et al. Evidence for a bidirectional relationship
between cytomegalovirus replication and acute graft-versus-host disease. Biol Blood Marrow Transplant. 2010;16:1309–14. https://doi.org/10.1016/j.bbmt.2010.03.020. Article PubMed Google
Scholar Download references ACKNOWLEDGEMENTS We would like to thank Prof. Robert Zeiser (University of Freiburg/Germany) for critical reading of this manuscript. We thank Martin Verniquet
for critical review of statistical analyses. We would also like to thank Nicole Raus (SFGM-TC, Lyon, France) for retrieving the clinical data from the ProMISe database. This work was
supported by grants from the Agence Nationale de la Recherche (ANR) (ANR-11-LABX-0070_TRANSPLANTEX), and the INSERM (UMR_S 1109), the Institut Universitaire de France (IUF), all to SB; from
the University of Strasbourg (IDEX UNISTRA) to CP and SB; from the European regional development fund (European Union) INTERREG V program (project n°3.2 TRIDIAG) to RC and SB; and from
MSD-Avenir grant AUTOGEN to SB. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Laboratoire d’ImmunoRhumatologie Moléculaire, INSERM UMR_S1109, Plateforme GENOMAX, Faculté de Médecine,
Fédération Hospitalo-Universitaire OMICARE, Fédération de Médecine Translationnelle de Strasbourg (FMTS), Université de Strasbourg, Strasbourg, France Raphael Carapito, Ismail Aouadi,
Angélique Pichot, Perrine Spinnhirny, Cécile Macquin, Véronique Rolli, Catherine Paillard, Dominique Charron & Seiamak Bahram * Labex TRANSPLANTEX, Faculté de Médecine, Université de
Strasbourg, Strasbourg, France Raphael Carapito, Ismail Aouadi, Angélique Pichot, Perrine Spinnhirny, Aurore Morlon, Irina Kotova, Cécile Macquin, Véronique Rolli, Anne Cesbron, Katia Gagne,
Pascale Loiseau, Ryad Tamouza, Antoine Toubert, Catherine Paillard, Régis Peffault de Latour, Dominique Charron, Gérard Socié & Seiamak Bahram * INSERM Franco-Japanese Nextgen HLA
Laboratory, Strasbourg, France Raphael Carapito, Ismail Aouadi, Angélique Pichot, Perrine Spinnhirny, Cécile Macquin, Véronique Rolli & Seiamak Bahram * INSERM Franco-Japanese Nextgen
HLA Laboratory, Nagano, Japan Raphael Carapito, Ismail Aouadi, Angélique Pichot, Perrine Spinnhirny, Cécile Macquin, Véronique Rolli & Seiamak Bahram * Laboratoire d’Immunologie, Plateau
Technique de Biologie, Pôle de Biologie, Nouvel Hôpital Civil, Strasbourg, France Raphael Carapito & Seiamak Bahram * BIOMICA SAS, Strasbourg, France Aurore Morlon & Irina Kotova *
Etablissement Français du Sang (EFS) Centre-Pays de la Loire, Laboratoire HLA, Nantes, France Anne Cesbron & Katia Gagne * Société Francophone de Greffe de Moelle et de Thérapie
Cellulaire (SFGM-TC), Hôpital Edouard Herriot, CHU, Lyon, France Anne Cesbron, Pascale Loiseau, Antoine Toubert, Anne Parissiadis, Catherine Paillard, Mauricette Michallet, Bruno Lioure,
Régis Peffault de Latour, Didier Blaise, Ibrahim Yakoub-Agha, Philippe Moreau, Mohamad Mohty & Gérard Socié * Société Francophone d’Histocompatibilité et d’Immunogénétique (SFHI), Paris,
France Anne Cesbron * INSERM 1232, CRCINA, Université Nantes-Angers, Nantes, France Katia Gagne * Europdonor operated by Matchis Foundation, Leiden, The Netherlands Machteld Oudshoorn *
Department of Immunohematology and Blood transfusion, LUMC, Leiden, The Netherlands Machteld Oudshoorn & Frans Claas * HOVON Data Center, Department of Hematology, Erasmus MC Cancer
Institute, Rotterdam, The Netherlands Bronno van der Holt * Laboratoire d’Immunologie, CHRU de Lille, Lille, France Myriam Labalette * LIRIC INSERM U995, Université Lille 2, Lille, France
Myriam Labalette & Ibrahim Yakoub-Agha * Laboratory for Translational Immunology, University Medical Center Utrecht, Utrecht, The Netherlands Eric Spierings * CNRS, EFS-PACA, ADES UMR
7268, Aix-Marseille Université, Marseille, France Christophe Picard * Laboratoire Jean Dausset, INSERM UMR_S 1160, Hôpital Saint-Louis, Paris, France Pascale Loiseau, Ryad Tamouza, Antoine
Toubert & Dominique Charron * Etablissement Français du Sang (EFS) Grand-Est, Laboratoire HLA, Strasbourg, France Anne Parissiadis * Etablissement Français du Sang (EFS) Rhône-Alpes,
Laboratoire HLA, Lyon, France Valérie Dubois * Service d’Hématologie et d’Oncologie pédiatrique, Hôpitaux Universitaires de Strasbourg, Strasbourg, France Catherine Paillard * Institut de
Recherche Mathématique Avancée, CNRS UMR 7501, LabEx Institut de Recherche en Mathématiques, ses Interactions et Applications, Université de Strasbourg, Strasbourg, France Myriam
Maumy-Bertrand & Frédéric Bertrand * Department of Hematology, Leiden University Medical Center, Leiden, The Netherlands Peter A. von dem Borne * Department of Hematology, University
Medical Center Utrecht, Utrecht, The Netherlands Jürgen H. E. Kuball * Centre Hospitalier Lyon Sud, Hématologie 1G, Hospices Civils de Lyon, Pierre Bénite, Lyon, France Mauricette Michallet
* Service d’Hématologie Adulte, Hôpitaux Universitaires de Strasbourg, Strasbourg, France Bruno Lioure & Gérard Socié * Service d’Hématologie – Greffe, Hôpital Saint-Louis, APHP, Paris,
France Régis Peffault de Latour * Institut Paoli Calmettes, Marseille, France Didier Blaise * Department of Hematology and ErasmusMC Cancer Institute, Erasmus University Medical Center,
Rotterdam, The Netherlands Jan J. Cornelissen * Service d’Hématologie Clinique, CHU Hôtel Dieu, Nantes, France Philippe Moreau * Département d’Hématologie, Hôpital Saint Antoine, Paris,
France Mohamad Mohty * Université Pierre & Marie Curie, Paris, France Mohamad Mohty * Centre de Recherche Saint-Antoine, INSERM UMR_S 938, Paris, France Mohamad Mohty * Division of
Epidemiology and Prevention, Aichi Cancer Center Research Institute, 1-1 Kanokoden, Chikusa-ku, Nagoya, Japan Yasuo Morishima Authors * Raphael Carapito View author publications You can also
search for this author inPubMed Google Scholar * Ismail Aouadi View author publications You can also search for this author inPubMed Google Scholar * Angélique Pichot View author
publications You can also search for this author inPubMed Google Scholar * Perrine Spinnhirny View author publications You can also search for this author inPubMed Google Scholar * Aurore
Morlon View author publications You can also search for this author inPubMed Google Scholar * Irina Kotova View author publications You can also search for this author inPubMed Google
Scholar * Cécile Macquin View author publications You can also search for this author inPubMed Google Scholar * Véronique Rolli View author publications You can also search for this author
inPubMed Google Scholar * Anne Cesbron View author publications You can also search for this author inPubMed Google Scholar * Katia Gagne View author publications You can also search for
this author inPubMed Google Scholar * Machteld Oudshoorn View author publications You can also search for this author inPubMed Google Scholar * Bronno van der Holt View author publications
You can also search for this author inPubMed Google Scholar * Myriam Labalette View author publications You can also search for this author inPubMed Google Scholar * Eric Spierings View
author publications You can also search for this author inPubMed Google Scholar * Christophe Picard View author publications You can also search for this author inPubMed Google Scholar *
Pascale Loiseau View author publications You can also search for this author inPubMed Google Scholar * Ryad Tamouza View author publications You can also search for this author inPubMed
Google Scholar * Antoine Toubert View author publications You can also search for this author inPubMed Google Scholar * Anne Parissiadis View author publications You can also search for this
author inPubMed Google Scholar * Valérie Dubois View author publications You can also search for this author inPubMed Google Scholar * Catherine Paillard View author publications You can
also search for this author inPubMed Google Scholar * Myriam Maumy-Bertrand View author publications You can also search for this author inPubMed Google Scholar * Frédéric Bertrand View
author publications You can also search for this author inPubMed Google Scholar * Peter A. von dem Borne View author publications You can also search for this author inPubMed Google Scholar
* Jürgen H. E. Kuball View author publications You can also search for this author inPubMed Google Scholar * Mauricette Michallet View author publications You can also search for this author
inPubMed Google Scholar * Bruno Lioure View author publications You can also search for this author inPubMed Google Scholar * Régis Peffault de Latour View author publications You can also
search for this author inPubMed Google Scholar * Didier Blaise View author publications You can also search for this author inPubMed Google Scholar * Jan J. Cornelissen View author
publications You can also search for this author inPubMed Google Scholar * Ibrahim Yakoub-Agha View author publications You can also search for this author inPubMed Google Scholar * Frans
Claas View author publications You can also search for this author inPubMed Google Scholar * Philippe Moreau View author publications You can also search for this author inPubMed Google
Scholar * Dominique Charron View author publications You can also search for this author inPubMed Google Scholar * Mohamad Mohty View author publications You can also search for this author
inPubMed Google Scholar * Yasuo Morishima View author publications You can also search for this author inPubMed Google Scholar * Gérard Socié View author publications You can also search for
this author inPubMed Google Scholar * Seiamak Bahram View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS RC performed the experiments,
designed the study, analyzed the data, and wrote the manuscript. SB designed the study, analyzed the data and wrote the manuscript. PS, AM, IK, CM, APi, and VR performed the experiments and
analyzed the data. IA performed the statistics. PAB, DB, AC, DC, FC, KG, JK, JC, ML, PL, MMi, PM, MOu, APa, RPL, CPi, GS, ES, RT, AT, IY, VD and BH provided samples and clinical data,
interpreted the clinical data and discussed the results. BL, MMo, AN, YM and CPa interpreted the clinical data and discussed results. FB, MMB, and BH analyzed the data and reviewed
statistics. All authors contributed to the writing of the report and approved the final version of the manuscript. CORRESPONDING AUTHORS Correspondence to Raphael Carapito or Seiamak Bahram.
ETHICS DECLARATIONS CONFLICT OF INTEREST SB is the scientific founder and a (minority) shareholder of BIOMICA SAS. JK is the co-founder and chief scientific officer of Gadeta. He received
personal fees from Gadeta. In addition, JK has a patent issued/pending. ES is the inventor of a patent application filed by the University Medical Center Utrecht on the prediction of an
alloimmune response against mismatched HLA (PCT/EPT2013/073386). All other authors declare no conflict of interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral
with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TABLE 1 FIGURE S1 FIGURE S1. LEGEND RIGHTS AND PERMISSIONS
Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Carapito, R., Aouadi, I., Pichot, A. _et al._ Compatibility at amino acid position 98 of MICB reduces the incidence of
graft-versus-host disease in conjunction with the CMV status. _Bone Marrow Transplant_ 55, 1367–1378 (2020). https://doi.org/10.1038/s41409-020-0886-5 Download citation * Received: 17
December 2019 * Revised: 17 March 2020 * Accepted: 23 March 2020 * Published: 14 April 2020 * Issue Date: July 2020 * DOI: https://doi.org/10.1038/s41409-020-0886-5 SHARE THIS ARTICLE Anyone
you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by
the Springer Nature SharedIt content-sharing initiative