Play all audios:
ABSTRACT BACKGROUND Analysis of long noncoding RNA (lncRNA) localisation at both the tissue and subcellular levels can provide important insights into the cell types that are important for
their function. METHODS By applying new fluorescent in situ hybridisation technique called hybridisation chain reaction (HCR), we achieved a high-throughput lncRNA visualisation and
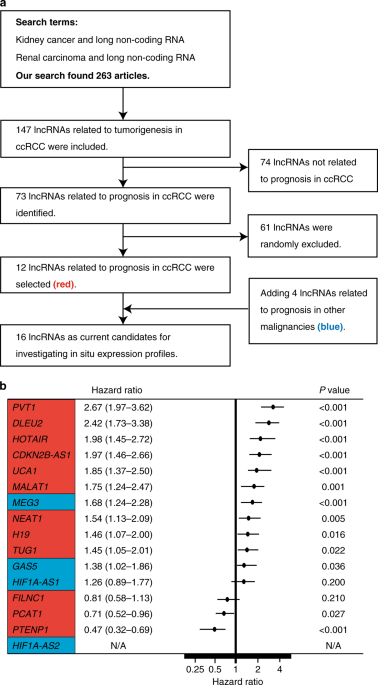
evaluation of clinical samples. RESULTS Assessing 1728 pairs of 16 lncRNAs and clear-cell renal-cell carcinoma (ccRCC) specimens, three lncRNAs (_TUG1_, _HOTAIR_ and _CDKN2B-AS1_) were
associated with ccRCC prognosis. Furthermore, we derived a new lncRNA risk group of ccRCC prognosis by combining the expression levels of these three lncRNAs. Examining genomic alterations
underlying this classification revealed prominent features of tumours that could serve as potential biomarkers for targeting lncRNAs. We then derived combination of HCR with expansion
microscopy and visualised nanoscale-resolution HCR signals in cell nuclei, uncovering intracellular colocalization of three lncRNA (_TUG1_, _HOTAIR_ and _CDKN2B-AS1_) signals such as those
located intra- or out of the nucleus or nucleolus in cancer cells. CONCLUSION LncRNAs are expected to be desirable noncoding targets for cancer diagnosis or treatments. HCR involves plural
probes consisting of small DNA oligonucleotides, clinically enabling us to detect cancerous lncRNA signals simply and rapidly at a lower cost. Access through your institution Buy or
subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 24 print issues and online
access $259.00 per year only $10.79 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which
are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS
EPIGENOMIC CHARTING AND FUNCTIONAL ANNOTATION OF RISK LOCI IN RENAL CELL CARCINOMA Article Open access 21 January 2023 _TRIM63_ IS A SENSITIVE AND SPECIFIC BIOMARKER FOR MIT FAMILY
ABERRATION-ASSOCIATED RENAL CELL CARCINOMA Article 14 April 2021 PIRNAS AND CIRCRNAS ACTING AS DIAGNOSTIC BIOMARKERS IN CLEAR CELL RENAL CELL CARCINOMA Article Open access 05 March 2025 DATA
AVAILABILITY All data supporting the findings of this study are included within the article and its Supplementary Information files (and Reporting summary). Also, the data will be shared
upon reasonable request to the corresponding author from colleagues who want to analyse in deep our findings. REFERENCES * Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et
al. Landscape of transcription in human cells. Nature. 2012;489:101–8. Article CAS Google Scholar * Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of
long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. Article CAS Google Scholar * Chu C, Spitale RC, Chang HY. Technologies to probe functions and mechanisms of
long noncoding RNAs. Nat Struct Mol Biol. 2015;22:29–35. Article CAS Google Scholar * Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–63. Article
CAS Google Scholar * Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, et al. Considerations when investigating lncRNA function in vivo. eLife. 2014;3:e03058. Article
Google Scholar * Choi HM, Chang JY, Trinh le A, Padilla JE, Fraser SE, Pierce NA. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat Biotechnol.
2010;28:1208–12. Article CAS Google Scholar * Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, et al. Third-generation in situ hybridization chain reaction:
multiplexed, quantitative, sensitive, versatile, robust. Development. 2018;145:dev165753. * Choi HM, Beck VA, Pierce NA. Next-generation in situ hybridization chain reaction: higher gain,
lower cost, greater durability. ACS Nano. 2014;8:4284–94. Article CAS Google Scholar * Sylwestrak EL, Rajasethupathy P, Wright MA, Jaffe A, Deisseroth K. Multiplexed intact-tissue
transcriptional analysis at cellular resolution. Cell. 2016;164:792–804. Article CAS Google Scholar * Shen H, Luo G, Chen Q. Long noncoding RNAs as tumorigenic factors and therapeutic
targets for renal cell carcinoma. Cancer Cell Int. 2021;21:110. Article CAS Google Scholar * Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell
Biol. 2021;220:e202009045. * Asano SM, Gao R, Wassie AT, Tillberg PW, Chen F, Boyden ES. Expansion microscopy: protocols for imaging proteins and RNA in cells and tissues. Curr Protoc Cell
Biol. 2018;80:e56. Article Google Scholar * Watanabe K, Kosaka T, Aimono E, Hongo H, Mikami S, Nishihara H, et al. Japanese case of enzalutamide-resistant prostate cancer harboring a SPOP
mutation with scattered allelic imbalance: response to platinum-based therapy. Clin Genitourin Cancer. 2019;17:e897–e902. Article Google Scholar * Takamatsu K, Tanaka N, Hakozaki K,
Takahashi R, Teranishi Y, Murakami T, et al. Profiling the inhibitory receptors LAG-3, TIM-3, and TIGIT in renal cell carcinoma reveals malignancy. Nat Commun. 2021;12:5547. Article CAS
Google Scholar * Győrffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Computational Struct Biotechnol J.
2021;19:4101–9. Article Google Scholar * Tumkur Sitaram R, Landström M, Roos G, Ljungberg B. Significance of PI3K signalling pathway in clear cell renal cell carcinoma in relation to VHL
and HIF status. J Clin Pathol. 2021;74:216–22. Article Google Scholar * D’Avella C, Abbosh P, Pal SK, Geynisman DM. Mutations in renal cell carcinoma. Urologic Oncol. 2020;38:763–73.
Article Google Scholar * Wang Y, Li Z, Li W, Zhou L, Jiang Y. Prognostic significance of long non-coding RNAs in clear cell renal cell carcinoma: a meta-analysis. Medicine 2019;98:e17276.
Article CAS Google Scholar * Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global
properties and specific subclasses. Genes Dev. 2011;25:1915–27. Article CAS Google Scholar * Polovic M, Dittmar S, Hennemeier I, Humpf HU, Seliger B, Fornara P, et al. Identification of a
novel lncRNA induced by the nephrotoxin ochratoxin A and expressed in human renal tumor tissue. Cell Mol Life Sci: CMLS. 2018;75:2241–56. Article CAS Google Scholar * Shi H, Sun Y, He M,
Yang X, Hamada M, Fukunaga T, et al. Targeting the TR4 nuclear receptor-mediated lncTASR/AXL signaling with tretinoin increases the sunitinib sensitivity to better suppress the RCC
progression. Oncogene. 2020;39:530–45. Article CAS Google Scholar * Miranda-Castro R, de-Los-Santos-Álvarez N, Lobo-Castañón MJ. Long noncoding RNAs: from genomic junk to rising stars in
the early detection of cancer. Anal Bioanal Chem. 2019;411:4265–75. Article CAS Google Scholar * Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future
perspectives. Mol Cancer. 2016;15:39. Article Google Scholar * Katayama H, Tamai K, Shibuya R, Nakamura M, Mochizuki M, Yamaguchi K, et al. Long non-coding RNA HOTAIR promotes cell
migration by upregulating insulin growth factor-binding protein 2 in renal cell carcinoma. Sci Rep. 2017;7:12016. Article Google Scholar * Wang PQ, Wu YX, Zhong XD, Liu B, Qiao G.
Prognostic significance of overexpressed long non-coding RNA TUG1 in patients with clear cell renal cell carcinoma. Eur Rev Med Pharmacol Sci. 2017;21:82–6. PubMed Google Scholar * Xie X,
Lin J, Fan X, Zhong Y, Chen Y, Liu K, et al. LncRNA CDKN2B-AS1 stabilized by IGF2BP3 drives the malignancy of renal clear cell carcinoma through epigenetically activating NUF2 transcription.
Cell Death Dis. 2021;12:201. Article CAS Google Scholar * Zhang K, Shi ZM, Chang YN, Hu ZM, Qi HX, Hong W. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene.
2014;547:1–9. Article CAS Google Scholar * Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic
Acids Res. 2016;44:863–77. Article CAS Google Scholar * Goldfarb KC, Cech TR. Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing. Genes Dev.
2017;31:59–71. Article CAS Google Scholar * Noh JH, Kim KM, Abdelmohsen K, Yoon JH, Panda AC, Munk R, et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of
the lncRNA RMRP. Genes Dev. 2016;30:1224–39. Article CAS Google Scholar Download references FUNDING This study was supported by Grants-in-Aid for Scientific Research (KAKENHI 19K18598 and
21K09356 to RK; 19H03792, 21K19414, and 22H03217 to NT; and 18H02939 to MO) and grants from the Kobayashi Foundation for Cancer Research (to NT), the SGH Foundation for Cancer Research (to
NT), the JUA Research Grant (to NT), the Princess Takamatsu Cancer Research Fund (to NT), and the Keio Gijuku Academic Development Funds (to NT). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS
* Department of Urology, Keio University School of Medicine, 160-8582, Tokyo, Japan Ryohei Kufukihara, Nobuyuki Tanaka, Kimiharu Takamatsu, Naoya Niwa, Keishiro Fukumoto, Yota Yasumizu,
Toshikazu Takeda, Kazuhiro Matsumoto, Shinya Morita, Takeo Kosaka, Ryuichi Mizuno & Mototsugu Oya * Genomics Unit, Keio Cancer Center, Keio University School of Medicine, Tokyo, Japan
Eriko Aimono & Hiroshi Nishihara Authors * Ryohei Kufukihara View author publications You can also search for this author inPubMed Google Scholar * Nobuyuki Tanaka View author
publications You can also search for this author inPubMed Google Scholar * Kimiharu Takamatsu View author publications You can also search for this author inPubMed Google Scholar * Naoya
Niwa View author publications You can also search for this author inPubMed Google Scholar * Keishiro Fukumoto View author publications You can also search for this author inPubMed Google
Scholar * Yota Yasumizu View author publications You can also search for this author inPubMed Google Scholar * Toshikazu Takeda View author publications You can also search for this author
inPubMed Google Scholar * Kazuhiro Matsumoto View author publications You can also search for this author inPubMed Google Scholar * Shinya Morita View author publications You can also search
for this author inPubMed Google Scholar * Takeo Kosaka View author publications You can also search for this author inPubMed Google Scholar * Eriko Aimono View author publications You can
also search for this author inPubMed Google Scholar * Hiroshi Nishihara View author publications You can also search for this author inPubMed Google Scholar * Ryuichi Mizuno View author
publications You can also search for this author inPubMed Google Scholar * Mototsugu Oya View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS
RK, NT and MO designed the study. RK, KT and EA performed the experiments. YY, TT, KM, SM, TK, HN and RM provided conceptual advice. RK and NT wrote the manuscript. CORRESPONDING AUTHOR
Correspondence to Nobuyuki Tanaka. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ETHICS APPROVAL AND CONSENT TO PARTICIPATE All procedures were
performed in approval of the Research Ethics Committee of Keio University (Approval Nos.: 20180098 and 20190059) and in compliance with the 1964 Helsinki Declaration and present ethical
standards. Both written informed consent and passive (opt-out) informed consent procedures have been applied to the experimental use of human samples. Opt-out informed consent from patients
was obtained by exhibiting the research information on our department's website (Department of Urology, Keio University Hospital, Tokyo, Japan). The need to obtain written informed
consent was waived if patients had finished their follow-up or had died, due to the study’s observational nature and the urgent need for cancer patient care. This was approved and reviewed
by the Research Ethics Committee of Keio University, in accordance with the ethical guidelines for Medical and Health Research Involving Human Subjects (Public Notice of the Ministry of
Education, Culture, Sports, Science and Technology and the Ministry of Health, Labor and Welfare as of July 2018; https://www.lifescience.mext.go.jp/files/pdf/n2181_01.pdf). CONSENT TO
PUBLISH Not applicable. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION REPORTING SUMMARY RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Kufukihara, R., Tanaka, N.,
Takamatsu, K. _et al._ Hybridisation chain reaction-based visualisation and screening for lncRNA profiles in clear-cell renal-cell carcinoma. _Br J Cancer_ 127, 1133–1141 (2022).
https://doi.org/10.1038/s41416-022-01895-3 Download citation * Received: 10 November 2021 * Revised: 03 June 2022 * Accepted: 09 June 2022 * Published: 28 June 2022 * Issue Date: 05 October
2022 * DOI: https://doi.org/10.1038/s41416-022-01895-3 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable
link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative