Play all audios:
ABSTRACT Heparan sulfate proteoglycan 2 (HSPG2), also known as perlecan, is a large multi-domain extracellular matrix proteoglycan, which contributes to the invasion, metastasis and
angiogenesis of solid tumor. However, very little is known about the effect of HSPG2 on acute myeloid leukemia (AML). This study aims to investigate the prognostic value of the HSPG2 gene in
terms of overall survival and leukemia-free survival in patients with AML. Bone marrow mononuclear cells (BMMCs) from 4 AML patients and 3 healthy controls were processed for RNA-Sequencing
(RNA-seq). The mRNA expression level of HSPG2 in BMMCs and CD34+ hematopoietic stem/progenitor cells (HSPC) obtained from enrolled participants and human leukemic cell lines was detected by
RT-qPCR. Then the correlations between the expression of HSPG2 and a variety of important clinical parameters, such as median white blood cell (WBC) count and bone marrow (BM) blasts, were
further analyzed. The expression level of HSPG2 was significantly upregulated in AML patients at the time of diagnosis, downregulated after complete remission and then elevated again at
relapse. Moreover, HSPG2 expression was associated with median WBC count (_P_ < 0.001), median hemoglobin (_P_ = 0.02), median platelet count (_P_ = 0.001), and BM blasts (_P_ < 0.001)
in AML patients. Patients with high HSPG2 expression had both worse overall survival (OS) (_P_ = 0.001) and poorer leukemia-free survival (LFS) (_P_ = 0.047). In the multivariate analysis
model, HSPG2 was identified as an independent prognostic biomarker of AML. Taken together, these results indicate that HSPG2 overexpression was associated with poor prognosis in AML
patients, and may be a prognostic biomarker and therapeutic target of AML. SIMILAR CONTENT BEING VIEWED BY OTHERS GLOBAL MICRORNA PROFILING OF BONE MARROW-MSC DERIVED EXTRACELLULAR VESICLES
IDENTIFIES MIRNAS ASSOCIATED WITH HEMATOPOIETIC DYSFUNCTION IN APLASTIC ANEMIA Article Open access 23 August 2024 A BIOINFORMATICS ANALYSIS AND EXPERIMENTAL VALIDATION OF PDGFD AS A
PROMISING DIAGNOSTIC BIOMARKER FOR ACUTE MYELOID LEUKEMIA Article Open access 28 April 2025 SINGLE-CELL ANALYSIS REVEALS ALTERED TUMOR MICROENVIRONMENTS OF RELAPSE- AND REMISSION-ASSOCIATED
PEDIATRIC ACUTE MYELOID LEUKEMIA Article Open access 05 October 2023 INTRODUCTION Acute myeloid leukemia (AML) is a group of malignant myelopoietic stem/progenitor cell diseases,
characterized by abnormal proliferation of primitive and immature myeloid cells in bone marrow and peripheral blood1,2. Genetic abnormalities, including chromosomal abnormalities, gene
mutations and gene expression abnormalities, involved in the pathogenesis of AML. For example, mutations in NPM1, FLT3-ITD, and c-KIT, as well as aberrant expression of BCL-2, MN1, WT1, and
MDR1 provide some clues for evaluating the prognosis of AML patients3,4,5. Although most AML patients achieve complete remission (CR) after chemotherapy and allogeneic hematopoietic stem
cell transplantation (allo-HSCT), the 5-year overall survival rate of AML patients is still poor6. Drug resistance and relapse are the two main reasons that affect the survival of patients
after transplantation. Therefore, it is of great clinical significance to identify new biomarkers for predicting prognosis and optimizing individual treatment strategies. HSPG2 (Perlecan), a
heparan sulfate proteoglycan, is a protein encoded by HSPG2 gene (maps to 1p36.12 in the human genome)7,8,9. 1p36 is a Carcinoma Prostate Brain (CAPB) locus that hosts a series of oncogenes
for prostate cancer and the brain tumor10. Functionally, HSPG2 is one of the major components of the vascular extracellular matrix and basement membranes11. Previous studies have showed
that high expression of HSPG2 might be an indicator of prostate cancer grade, invasion potential and distant metastasis10,12. Besides, it is reported that the overexpression of HSPG2
predicts poor survival in patients with oligoastrocytoma and oligodendroglioma13,14. An in vitro study found that HSPG2 is actively synthesized by bone marrow derived cells and the abnormal
expression of HSPG2 might play a vital role in hematopoietic cell differentiation15. However, the role of HSPG2 remains unknown in AML. In this study, RNA-sequencing (RNA-seq) and
differential expression analysis found that the overexpression of HSPG2 was higher in AML patients significantly than in healthy controls. The role of HSPG2 remains unknown in AML, although
overexpression of HSPG2 has been well studied in other diseases. Our research aimed to detect the expression of HSPG2 in AML patients and assess the influence of HSPG2 expression on the
clinical characteristics and prognosis, which might contribute to explore new biomarker and therapeutic target of AML. MATERIALS AND METHODS PATIENTS AND CLINICAL CHARACTERISTICS Bone marrow
specimens and clinical data were obtained from patients who were diagnosed with AML between 2014 and 2017. In total 151 AML patients and 11 healthy controls were enrolled in this study. The
use of bone marrow specimens was approved by the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (2020–264). Written informed consent was obtained from all
participants. Bone marrow mononuclear cells (BMMCs) from the participants were separated by density-gradient centrifugation with Lymphocyte Separation Medium (TBD, China, LTS1077). The main
clinical and laboratory features of the patients were presented in Table 2. CD34+ CELL SORTING Normal bone marrow CD34+ cells and AML bone marrow CD34+ cells were isolated from BMMCs by
using EasySepTM Human CD34 Positive Selection Kit II (Stemcell Technologies, Canada). Briefly, 1 × 108 cells were incubated with 20 μL Selection Cocktail for 30 min at room temperature.
Then, the cells were incubated in a magnet with 15 μL RapidSphereTM for 5 min at room temperature. After pouring off the supernatant, cells were resuspended in 1 mL of PBS to pick up the
magnet coupled with CD34+ cells in five times in order to yield a high purity (>95%). Finally, the purity of collected CD34+ cells was estimated by flow cytometry (FACSCalibur analyzer,
BD Biosciences, USA). CELL LINES AND CELL CULTURE The SKM-1, an AML-MDS cell line, was provided by Professor Jianfeng Zhou working in Tongji Medical College of Huazhong University of Science
and Technology (Wuhan, China). Chronic myeloid leukemia cell line K562 was provided by Chongqing Key Laboratory of Translational Medicine in Major Metabolic Diseases. The HS-5, a human bone
marrow stromal cell line, was provided by Department Molecular Diagnostic Center for Clinical Medicine, the First Affiliated Hospital of Chongqing Medical University (Chongqing, China).
Human peripheral blood leukemia T cells line Jurkat was provided by Children’s Hospital of Chongqing Medical University (Chongqing, China). HS-5 cells were cultured in DMEM medium
(Gibco/Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (FBS, PAN seratech, Germany) and 1% penicillin/streptomycin (PS, Beyotime, China). Three human leukemia cell
lines (SKM-1, K562, and Jurkat) were cultured in RPMI 1640 medium (Gibco/Thermo Fisher Scientific, USA) supplemented with 10% FBS (PAN seratech, Germany) and 1% PS (Beyotime, China). The
cells were cultured in a humid atmosphere at 37 °C with 5% CO2. RNA ISOLATION AND QUANTITATIVE REAL-TIME POLYMERASE CHAIN REACTION (RT-QPCR) Total RNA was isolated from each sample using
trizol reagent (Beyotime, China). Complementary DNA (cDNA) was synthesized from total RNA with reverse transcription kit (Takara, Japan) according to the manufacturer’s instructions. RT-qPCR
was performed using CFX96 Real-Time PCR Detection System (BIO RAD, USA). The total reaction volume was 10 µL and was prepared as follows: 5 µL of TB Green (Takara, Japan), 0.4 µL of each
primer (10 µmol/L), 1 µL of cDNA template (0.5 ng/µL), and 3.2 µL of ddH2O. The cycling conditions were as follows: 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s and 60 °C for 30 s.
Transcript levels were normalized vs. β-actin expression. The gene expression was calculated using the formula 2−ΔΔCt. The primers sequences were as follows: HSPG2 Forward
5’-GACATCGCCATGGATACCAC-3’ Reverse 5’-CAGGACAAGCCAGAATAGCC-3’ β-actin Forward 5’-CATTGCCGACAGGATGCAG-3’ Reverse 5’-CGGAGTACTTGCGCTCAGGA-3’. WESTERN BLOT Total protein was harvested from each
sample using RIPA lysis buffer (Beyotime, China) supplemented with 1 µM phenylmethanesulfonyl fluoride (PMSF, Beyotime, China). Then, the extract was digested with 0.01 units/ml heparinase
III (Sigma, USA, H8891) at 37 °C for 3 h. Thirty microgram of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, GenScript, USA) and transferred
onto polyvinylidene fluoride membranes (PVDF, Millipore, USA). Subsequently, membranes were blocked with 5% non-fat milk for 2 h at room temperature, followed by incubation with primary
antibodies at 4 °C overnight. The primary antibodies used in this study were anti-HSPG2 (SantaCruz, USA, sc-33707) used at 1:1000 and anti-β-actin (Bioss, China, bs-0061R) used at 1:5000
diluted in primary antibody dilution buffer (Beyotime, China, P0256). The membranes were washed and exposed to corresponding horseradish peroxidase (HRP)-conjugated goat anti-rat (1:5000,
Mengbio, China, MS002A) or goat anti-rabbit (1:1000, Beyotime, China, A0208) secondary antibodies diluted in TBST buffer for 1 h at room temperature. Finally, the protein bands were
visualized with an enhanced chemiluminescence (ECL) kit (Advansta, USA, K-12045-D10), and the band intensity was analyzed using Vilber Fusion software (Fusion FX5 Spectra, France). RNA-SEQ
Paired-end libraries were synthesized by using the TruSeq™ RNA Sample Preparation Kit (Illumina, USA) following TruSeq™ RNA Sample Preparation Guide. Briefly, the poly-A containing mRNA
molecules were purified using poly-T oligo-attached magnetic beads. Following purification, the mRNA is fragmented into small pieces. The cleaved RNA fragments are copied into first strand
cDNA. This is followed by second strand cDNA synthesis using DNA Polymerase I and RNase H. These cDNA fragments then go through an end repair process, the addition of a single ‘A’ base, and
then ligation of the adapters. The products are then purified and enriched with PCR to create the final cDNA library. Cluster was generated by cBot with the library diluted to 10 pM and then
was sequenced on the Illumina NovaSeq 6000 (Illumina, USA). The library construction and sequencing were performed at Shanghai Sinomics Corporation, followed by data analysis they
performed. The criteria for differential genes was set up as fold change >2 or <−2, _P_-value <0.05, and FDR < 0.05. STATISTICAL ANALYSIS Data were analyzed using SPSS 22.0 (SPSS
Inc., USA) and GraphPad Prism 5.01 (GraphPad Software Inc., USA). Complete remission (CR) was defined as normalization of peripheral blood and bone marrow (<5% blasts in the bone marrow;
absence of extramedullary disease; absolute neutrophil count >1.0 × 109/L; platelet count >100 × 109/L), and absence of clinical symptoms16. Overall survival (OS) was measured from
diagnosis to last follow-up or death from any cause. Leukemia-free survival (LFS) was calculated from the day that CR was established until either relapse or death without relapse17.
Mann–Whitney’s U test and Pearson Chi-square analysis/Fisher exact test were applied to compare the difference of continuous variables and categorical variables between two groups,
respectively. The ROC curve and area under the ROC curve (AUC) were to assess the discriminative capacity of HSPG2 expression among patients and controls. The univariate Kaplan–Meier (KM)
method and the multivariate Cox proportional hazard regression model were used to determine the survival curve and independent risk factors of AML patients. Spearman was used to analyze the
correlation between the expression level of HSPG2 and clinicopathological factors. For all tests, a _P_-value < 0.05 was considered as statistically significant. All experiments were
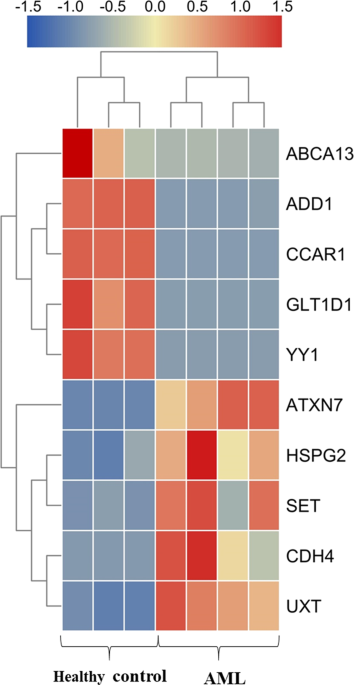
performed in triplicate. RESULTS THE EXPRESSION OF HSPG2 IN PARTICIPANTS AND CELL LINES The results of RNA-seq analysis indicated HSPG2 significantly upregulation in AML patients compared
with healthy controls (Fig. 1 and Table 1). As shown in the results of RT-qPCR, the expression of HSPG2 was significantly upregulated in AML patients (Fig. 2a, _P_ < 0.001), and human
myeloid leukemia cell lines (SKM-1 cells and K562 cells) (Fig. 2c, _P_ < 0.001), but not in human T cell acute lymphoblastic leukemia cell line (Jurkat cells) (Fig. 2c, _P_ > 0.05).
The mRNA expression of HSPG2 in CD34+/CD34− cells from AML patients was higher than that in CD34+/CD34− cells from healthy controls (Fig. S1). Furthermore, the protein expression of HSPG2
was higher in AML patients than that in healthy controls (Fig. 2d). DISCRIMINATIVE CAPACITY OF HSPG2 EXPRESSION ROC curve was used to examine the diagnostic value of HSPG2 in AML patients
compared with healthy controls and found the AUC value to be 0.903 in whole-cohort AML (95% confidence interval (CI): 0.842–0.965, _P_ < 0.001; Fig. 3a) with a sensitivity of 88.0% and a
specificity of 74.1%, which indicated that HSPG2 could be a potential biomarker for distinguishing AML patients from healthy controls. Similarly, distinguishing capacity of HSPG2 expression
was also shown in non-M3 AML and cytogenetically normal AML patients (AUC = 0.888, 95% CI: 0.814–0.963, _P_ < 0.001; AUC = 0.834, 95% CI: 0.719–0.949, _P_ = 0.001; respectively; Fig. 3b,
c). Furthermore, to determine whether HSPG2 could be used as a marker of disease prognosis, the predictive capacity of HSPG2 levels was analyzed among AML cases and compared to BM blast
count, an established prognostic marker for AML. As highlighted by the comparison of ROC curves (Fig. 3d), HSPG2 expression alone is enough to obtain high predictive accuracy for 2-year
survival in whole-cohort AML (HSPG2 expression-AUC = 0.663, 95% CI: 0.572–0.754, _P_ = 0.002), as compared with the predictive capacity of BM blasts (BM blasts-AUC = 0.613, 95% CI:
0.516–0.710, _P_ = 0.034). CORRELATION OF HSPG2 EXPRESSION WITH CLINICAL CHARACTERISTICS IN AML By the set cutoff value based on the ROC curve, the whole cohort of AML patients were divided
into two groups, low HSPG2 expression (≤4.39) (HSPG2low) and high HSPG2 expression (>4.39) (HSPG2high). The comparisons of clinical manifestations and laboratory features between two
groups were presented in Table 2. Interestingly, the high expression of HSPG2 was found to be associated with higher WBC (_P_ < 0.001), lower hemoglobin (_P_ = 0.002), lower platelets
(_P_ = 0.001), and higher BM blasts (_P_ < 0.001). There were no significant differences in sex, age, French-American-British classification, karyotypes and treatment regimen between
HSPG2low and HSPG2high patients. In addition, no significant correlations between HSPG2 expression and seven gene mutations were confirmed (_P_ > 0.05). HSPG2 EXPRESSION IN THE MONITORING
OF AML To investigate HSPG2 expression in the monitoring of AML, we detected HSPG2 expression in patients at different clinical stages (151 patients at diagnosis, 41 patients at the time of
CR and 14 patients at the time of relapse). Notably, the expression level of HSPG2 was remarkably decreased in CR phase and then increased again in relapse phase (Fig. 2b). Moreover, CR
rate was also significantly higher in HSPG2low patients than in HSPG2high patients (_P_ = 0.011) (Table 2). CORRELATION BETWEEN HSPG2 EXPRESSION AND CLINICAL OUTCOME In this study, the
median follow-up period was 9 months. Kaplan–Meier survival analysis manifested that OS and LFS of patients with high HSPG2 expression were significantly shorter than those of patients with
low HSPG2 expression in the whole cohort of AML patients (_P_ < 0.001, Fig. 4a; _P_ = 0.047, Fig. 4d). Higher HSPG2 expression implied poor prognosis, which was also observed in the
Non-M3 cohort (OS: _P_ < 0.001, Fig. 4b; LFS: _P_ = 0.025, Fig. 4e) and CN-AML patients (OS: _P_ = 0.039, Fig. 4c; LFS: _P_ = 0.056, Fig. 4f). Univariate analyses and multivariate
analyses on OS and LFS in the whole-cohort AML patients were performed, including the expression level of HSPG2, age (≥60 vs. <60 years old), WBC (≥30 × 109/L vs. <30 × 109/L), BM
blasts(≥70% vs. <70%), treatment regimen and gene mutations (mutant vs. wild-type). As shown in Table 3, HSPG2 expression was significantly and independently associated with a worse OS
both in univariate (_P_ < 0.001) and multivariate analysis (_P_ = 0.004). Moreover, HSPG2 expression was associated with a poorer LFS in univariate analysis (_P_ = 0.047). Intriguingly,
as shown in the results of Spearman’s rank correlation analysis (Fig. 5a, b), there was a positive correlation between HSPG2 expression and BM blasts (_r_ = 0.486, _P_ < 0.001), as well
as WBC count (_r_ = 0.314, _P_ < 0.001). DISCUSSION Great progress has been obtained in our understanding of the pathogenesis of AML via the characterization of dysregulated genes, such
as transcription factor SALL418, ABC subfamily B-member 1 (ABCB1)19, and Homeodomain-only protein homeobox (HOPX)20. However, owing to the absence of an observation of the overall process
before and after disease progression of AML, those known differential expression genes are still unable to illuminate the mechanism underlying disease progression and prognosis. The factors
involved in disease progression and pathogenesis in AML remain to be elucidated. The five genes with a high upregulation and the five downregulation genes from our results of RNA-seq,
according to their relevance to the pathogenesis and prognosis of cancers were selected for further investigation. ABCA13 has identified as a marker of poor survival in metastatic ovarian
serous carcinoma21 and was found to be highly overexpressed in our results. ADD1 has been associated with primary hypertension22 and was also highly overexpressed. CCAR1 is a transcriptional
coactivator for nuclear receptors and exerts its functions as a key intracellular signal transducer of apoptosis signaling pathways23, which was overexpressed in AML patients in this study.
GLT1D1 was shown to be highly upregulated and promoted immunosuppression and tumor growth in B-cell non-Hodgkin’s lymphoma24. YY1 could be regulated by CXCR4 to mediate transcriptional
activation of MYC and BCLXL in AML cells25. ATXN7 was found to decrease in AML patients in our cohort, and low expression of ATXN7 was significantly associated with poor recurrence-free
survival (RFS) and OS in Hepatitis B Virus-Related Hepatocellular Carcinoma26. SET protein regulated intracellular redox state and sustained autophagy in Head and Neck Squamous Cell
Carcinoma (HNSCC) cells, which may play an important role in resistance to the death of HNSCC cells27. The expression of CDH4 was decreased in nasopharyngeal carcinoma cell lines, xenografts
and primary tumor biopsies and may be a novel putative tumor suppressor gene28. UXT is a novel regulator of the polycomb repressive complex 2 (PRC2) and acts as a renal cancer oncogene that
affects the progression and survival of clear cell renal cell carcinoma patients29. Moreover, the dysregulation expression of HSPG2 was shown to affect the growth, invasion, metastasis, and
angiogenesis of several tumors30,31,32. In this study, we found that HSPG2 expression was elevated significantly in newly diagnosed AML patients compared with healthy controls, which was
consistent with the results of RNA-seq. HSPG2 exists in various tissues and exerts effects on many biological processes33. The abnormal expression of HSPG2 has been reported in some
carcinomas. In accordance with our findings, previous studies have found mRNA level of HSPG2 was markedly increased in solid tumor tissue compared with that in normal tissue, such as
metastatic melanomas34, oral squamous carcinomas35 and hepatocellular carcinomas36. Particularly, our data showed that HSPG2 expression decreased after complete remission and returned during
relapse phase. Consequently, it is considered that HSPG2 can play a vital role in disease progression and be a new molecular maker in AML. It is noteworthy that HSPG2 is significantly
highly expressed in CD34+ cells of AML patients, suggesting that perlecan is indeed secreted by the hematopoietic stem/progenitor cells in AML, even though that perlecan is known to be
produced from mesenchymal stem cells37. This observation, combined with our data that the expression of HSPG2 was higher in human myeloid leukemia cell lines than in bone marrow stromal cell
line, explained why HSPG2 expression in AML patients was higher than that in healthy controls as mentioned above. Collectively, this study showed HSPG2 was significantly increased in AML
patients, which revealed that inhibition of HSPG2 is a potential therapeutic strategy in AML. In this study, the AUC value is preferable for HSPG2 expression to distinguish AML patients from
healthy controls, including CN-AML and non-M3 AML, revealing it might be a promising diagnostic biomarker. The correlation between HSPG2 expression and several clinical and laboratory
characteristics was then evaluated. One the one hand, high expression of HSPG2 was associated with lower hemoglobin and platelet count. On the other hand, high expression of HSPG2 was
associated with higher WBC and BM blast count, which exhibited a positive correlation. A previous study demonstrated that high WBC count is a prognostic factor in patients with acute myeloid
leukemia with the genotypic combination ‘NPMc (+) with FLT3-ITD’38. Guo RJ and coworkers39 also reported that high WBC count was an independent prognostic factor for a shorter OS and EFS.
It is reported that excess blasts are the strongest predictors for poor outcome of AML and associated with the disease progression40,41. This discovery, combined with our findings that the
ability of HSPG2 for predicting 2-year survival is similar to that of BM blasts in AML based on the AUC value and that there was a positive correlation between HSPG2 expression and BM
blasts, suggested that HSPG2 expression might be a prognostic factor for AML. It is well known that cytogenetic abnormalities and somatic mutations have been found at diagnosis in 20–70% of
AML patients and are critical in evaluating the outcome of AML patients42. Herein, we also explored whether the expression of HSPG2 was correlated with karyotypes and gene mutations.
Unfortunately, no significant correlation was observed, which may be due to limited sample size. In addition, we observed that CR rate was significantly lower in HSPG2high patients after
receiving induction chemotherapy, which indicated that HSPG2 expression might be related to the response to the treatment of AML. Furthermore, survival analysis revealed that patients with
high HSPG2 expression had a poor prognosis in whole cohort AML, non-M3 AML and CN-AML. We also found that HSPG2 was an independent prognostic factor for OS based on univariate and
multivariate analyses and for LFS based on univariate analysis. As reported in oligoastrocytoma and oligodendroglioma, high expression of HSPG2 could independently predict poor OS and RFS13.
Hence, HSPG2 expression could be used to predict inferior survival and assess treatment outcome in AML. It is worth mentioning the treatment regimen is a crucial factor for OS and LFS in
AML and we found that patients treated with hematopoietic stem cell transplantation (HSCT) had better OS and LFS compared with chemotherapy alone. Based on these results, we can draw the
conclusion that the expression level of HSPG2 could be used as an important indicator of prognosis in AML. Certainly, more in-depth studies on large-scale samples of AML patients are needed
to verify these findings. This study implied that the expression level of HSPG2 was upregulated in AML patients and correlated with adverse prognosis and disease progression in AML patients.
To sum up, HSPG2 potentially might act as a pro-oncogene in the pathogenesis of AML, and further functional researches should be conducted to confirm its role in vivo and in vitro.
CONCLUSION In summary, this is the first study to report the relationship of HSPG2 expression with the clinical outcomes in AML patients. This study confirmed that overexpression of HSPG2
was an adverse prognostic indicator for AML, although the mechanism underlying the role of HSPG2 in AML remains to be illustrated. REFERENCES * Short, N. J. et al. Advances in the treatment
of acute myeloid leukemia: new drugs and new challenges. _Cancer Discov._ 10, 506–525 (2020). PubMed Google Scholar * Dohner, H., Weisdorf, D. J. & Bloomfield, C. D. Acute myeloid
leukemia. _N. Engl. J. Med._ 373, 1136–1152 (2015). PubMed Google Scholar * Chen, J., Odenike, O. & Rowley, J. D. Leukaemogenesis: more than mutant genes. _Nat. Rev. Cancer_ 10, 23–36
(2010). CAS PubMed PubMed Central Google Scholar * Verhaak, R. G. & Valk, P. J. Genes predictive of outcome and novel molecular classification schemes in adult acute myeloid
leukemia. _Cancer Treat. Res._ 145, 67–83 (2010). PubMed Google Scholar * Handschuh, L. et al. Gene expression profiling of acute myeloid leukemia samples from adult patients with AML-M1
and -M2 through boutique microarrays, real-time PCR and droplet digital PCR. _Int. J. Oncol._ 52, 656–678 (2018). CAS PubMed Google Scholar * Estey, E. H. Acute myeloid leukemia: 2019
update on risk-stratification and management. _Am. J. Hematol._ 93, 1267–1291 (2018). PubMed Google Scholar * Farach-Carson, M. C. & Carson, D. D. Perlecan-a multifunctional
extracellular proteoglycan scaffold. _Glycobiology_ 17, 897–905 (2007). CAS PubMed Google Scholar * Lord, M. S. et al. The multifaceted roles of perlecan in fibrosis. _Matrix Biol._
68–69, 150–166 (2018). PubMed Google Scholar * Farach-Carson, M. C., Warren, C. R., Harrington, D. A. & Carson, D. D. Border patrol: insights into the unique role of perlecan/heparan
sulfate proteoglycan 2 at cell and tissue borders. _Matrix Biol._ 34, 64–79 (2014). CAS PubMed Google Scholar * Datta, M. W. et al. Perlecan, a candidate gene for the CAPB locus,
regulates prostate cancer cell growth via the Sonic Hedgehog pathway. _Mol. Cancer_ 5, 9 (2006). PubMed PubMed Central Google Scholar * Elgundi, Z. et al. Cancer metastasis: the role of
the extracellular matrix and the heparan sulfate proteoglycan perlecan. _Front. Oncol._ 9, 1482 (2019). PubMed Google Scholar * Grindel, B. J. et al. Matrilysin/matrix
metalloproteinase-7(MMP7) cleavage of perlecan/HSPG2 creates a molecular switch to alter prostate cancer cell behavior. _Matrix Biol._ 36, 64–76 (2014). CAS PubMed PubMed Central Google
Scholar * Ma, X. L. et al. Increased HSPG2 expression independently predicts poor survival in patients with oligoastrocytoma and oligodendroglioma. _Eur. Rev. Med. Pharm. Sci._ 22,
6853–6863 (2018). Google Scholar * Kazanskaya, G. M. et al. Heparan sulfate accumulation and perlecan/HSPG2 up-regulation in tumour tissue predict low relapse-free survival for patients
with glioblastoma. _Histochem. Cell Biol._ 149, 235–244 (2018). CAS PubMed Google Scholar * Grassel, S., Cohen, I. R., Murdoch, A. D., Eichstetter, I. & Iozzo, R. V. The proteoglycan
perlecan is expressed in the erythroleukemia cell line K562 and is upregulated by sodium butyrate and phorbol ester. _Mol. Cell Biochem._ 145, 61–68 (1995). CAS PubMed Google Scholar *
Zhu, K. W. et al. Association of genetic polymorphisms in genes involved in Ara-C and dNTP metabolism pathway with chemosensitivity and prognosis of adult acute myeloid leukemia (AML). _J.
Transl. Med._ 16, 90 (2018). CAS PubMed PubMed Central Google Scholar * Zhou, J. D. et al. Identification and validation of SRY-box containing gene family member SOX30 methylation as a
prognostic and predictive biomarker in myeloid malignancies. _Clin. Epigenet._ 10, 92 (2018). Google Scholar * Ma, Y. et al. SALL4, a novel oncogene, is constitutively expressed in human
acute myeloid leukemia (AML) and induces AML in transgenic mice. _Blood_ 108, 2726–2735 (2006). CAS PubMed PubMed Central Google Scholar * Boyer, T. et al. Clinical significance of ABCB1
in acute myeloid leukemia: a comprehensive study. _Cancers_ 11, 1323 (2019). CAS PubMed Central Google Scholar * Lin, C. C. et al. Higher HOPX expression is associated with distinct
clinical and biological features and predicts poor prognosis in de novo acute myeloid leukemia. _Haematologica_ 102, 1044–1053 (2017). CAS PubMed PubMed Central Google Scholar * Nymoen,
D. A., Holth, A., Hetland Falkenthal, T. E., Tropé, C. G. & Davidson, B. CIAPIN1 and ABCA13 are markers of poor survival in metastatic ovarian serous carcinoma. _Mol. Cancer_ 14, 44
(2015). PubMed PubMed Central Google Scholar * Jin, H., Huang, Y. & Yang, G. Association between α-adducin rs4961 polymorphism and hypertension: a meta-analysis based on 40 432
subjects. _J. Cell Biochem._ 120, 4613–4619 (2019). CAS PubMed Google Scholar * Ou, C. Y., Kim, J. H., Yang, C. K. & Stallcup, M. R. Requirement of cell cycle and apoptosis regulator
1 for target gene activation by Wnt and beta-catenin and for anchorage-independent growth of human colon carcinoma cells. _J. Biol. Chem._ 284, 20629–20637 (2009). CAS PubMed PubMed
Central Google Scholar * Liu, X. et al. Overexpression of GLT1D1 induces immunosuppression through glycosylation of PD-L1 and predicts poor prognosis in B-cell lymphoma. _Mol. Oncol._ 14,
1028–1044 (2020). PubMed PubMed Central Google Scholar * Chen, Y. et al. CXCR4 downregulation of let-7a drives chemoresistance in acute myeloid leukemia. _J. Clin. Invest._ 123, 2395–2407
(2013). CAS PubMed PubMed Central Google Scholar * Han, C. et al. ATXN7 gene variants and expression predict post-operative clinical outcomes in Hepatitis B virus-related hepatocellular
carcinoma. _Cell Physiol. Biochem._ 39, 2427–2438 (2016). CAS PubMed Google Scholar * Ouchida, A. T. et al. SET protein accumulation prevents cell death in head and neck squamous cell
carcinoma through regulation of redox state and autophagy. _Biochim. Biophys. Acta Mol. Cell Res._ 1866, 623–637 (2019). CAS PubMed Google Scholar * Du, C. et al. CDH4 as a novel putative
tumor suppressor gene epigenetically silenced by promoter hypermethylation in nasopharyngeal carcinoma. _Cancer Lett._ 309, 54–61 (2011). CAS PubMed Google Scholar * Zeng, J. et al.
Ubiquitous expressed transcript promotes tumorigenesis by acting as a positive modulator of the polycomb repressive complex 2 in clear cell renal cell carcinoma. _BMC Cancer_ 19, 874 (2019).
PubMed PubMed Central Google Scholar * Monami, G. et al. Proepithelin regulates prostate cancer cell biology by promoting cell growth, migration, and anchorage-independent growth. _Am.
J. Pathol._ 174, 1037–1047 (2009). CAS PubMed PubMed Central Google Scholar * Qiang, B. et al. Perlecan heparan sulfate proteoglycan is a critical determinant of angiogenesis in response
to mouse hind-limb ischemia. _Can. J. Cardiol._ 30, 1444–1451 (2014). PubMed Google Scholar * Jiang, X. et al. Essential contribution of tumor-derived perlecan to epidermal tumor growth
and angiogenesis. _J. Histochem. Cytochem._ 52, 1575–1590 (2004). CAS PubMed Google Scholar * Martinez, J. R., Dhawan, A. & Farach-Carson, M. C. Modular proteoglycan perlecan/HSPG2:
mutations, phenotypes, and functions. _Genes_ 9, 556 (2018). PubMed Central Google Scholar * Cohen, I. R. et al. Abnormal expression of perlecan proteoglycan in metastatic melanomas.
_Cancer Res._ 54, 5771–5774 (1994). CAS PubMed Google Scholar * Kawahara, R. et al. Agrin and perlecan mediate tumorigenic processes in oral squamous cell carcinoma. _PLoS ONE_ 9, e115004
(2014). PubMed PubMed Central Google Scholar * Carloni, V., Luong, T. V. & Rombouts, K. Hepatic stellate cells and extracellular matrix in hepatocellular carcinoma: more complicated
than ever. _Liver Int._ 34, 834–843 (2014). PubMed Google Scholar * Nakamura, R., Nakamura, F. & Fukunaga, S. Contrasting effect of perlecan on adipogenic and osteogenic
differentiation of mesenchymal stem cells in vitro. _Anim. Sci. J._ 85, 262–270 (2014). CAS PubMed Google Scholar * de Jonge, H. J. et al. Prognostic impact of white blood cell count in
intermediate risk acute myeloid leukemia: relevance of mutated NPM1 and FLT3-ITD. _Haematologica_ 96, 1310–1317 (2011). PubMed PubMed Central Google Scholar * Guo, R. J., Atenafu, E. G.,
Craddock, K. & Chang, H. Allogeneic hematopoietic cell transplantation may alleviate the negative prognostic impact of monosomal and complex karyotypes on patients with acute myeloid
leukemia. _Biol. Blood Marrow Transpl._ 20, 690–695 (2014). Google Scholar * Holtzman, N. G. et al. Peripheral blood blast rate of clearance is an independent predictor of clinical response
and outcomes in acute myeloid leukaemia. _Br. J. Haematol._ 188, 881–887 (2020). CAS PubMed Google Scholar * Ram, R. et al. Venetoclax in patients with acute myeloid leukemia refractory
to hypomethylating agents-a multicenter historical prospective study. _Ann. Hematol._ 98, 1927–1932 (2019). CAS PubMed Google Scholar * Bullinger, L., Dohner, K. & Dohner, H. Genomics
of acute myeloid leukemia diagnosis and pathways. _J. Clin. Oncol._ 35, 934–946 (2017). CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We thanked the Chongqing Key
Laboratory of Translational Medicine in Major Metabolic Diseases (The First Affiliated Hospital of Chongqing Medical University, Chongqing, China) for providing laboratory facilities. This
work was supported by Natural Science Foundation Project of Chongqing (cstc2018jcyjAX0688), Chongqing Science and Health joint project (2018ZDXM001), and Chongqing Education Commission
Foundation (KJ1702017). AUTHOR INFORMATION Author notes * These authors contributed equally: Xiaojia Zhou, Simin Liang AUTHORS AND AFFILIATIONS * Department of Hematology, The First
Affiliated Hospital of Chongqing Medical University, Chongqing, China Xiaojia Zhou, Simin Liang, Li Yang & Li Wang * The Center for Clinical Molecular Medical detection, The First
Affiliated Hospital of Chongqing Medical University, Chongqing, China Qian Zhan * Center for the Study of Hematological Malignancies, Karaiskakio Foundation, 2032, Nicosia, Cyprus Jianxiang
Chi Authors * Xiaojia Zhou View author publications You can also search for this author inPubMed Google Scholar * Simin Liang View author publications You can also search for this author
inPubMed Google Scholar * Qian Zhan View author publications You can also search for this author inPubMed Google Scholar * Li Yang View author publications You can also search for this
author inPubMed Google Scholar * Jianxiang Chi View author publications You can also search for this author inPubMed Google Scholar * Li Wang View author publications You can also search for
this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Li Wang. ETHICS DECLARATIONS CONFLICT OF INTEREST The authors declare that they have no conflict of interest.
ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Edited by T. Kaufmann
SUPPLEMENTARY INFORMATION TABLE S1 FIG. S1 RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use,
sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative
Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated
otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds
the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Zhou, X., Liang, S., Zhan, Q. _et al._ HSPG2 overexpression independently predicts poor survival in patients with acute myeloid leukemia.
_Cell Death Dis_ 11, 492 (2020). https://doi.org/10.1038/s41419-020-2694-7 Download citation * Received: 31 January 2020 * Revised: 13 June 2020 * Accepted: 16 June 2020 * Published: 30 June
2020 * DOI: https://doi.org/10.1038/s41419-020-2694-7 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable
link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative