Play all audios:
ABSTRACT _HCN1_ is one of four genes encoding hyperpolarization-activated cyclic nucleotide-gated channels. The phenotypic spectrum associated with _HCN1_ variants ranges from neonatal
developmental and epileptic encephalopathy to idiopathic generalized epilepsy. We report a Japanese patient with repetitive focal seizures and super-refractory status epilepticus since early
infancy caused by a _de novo HCN1_ variant, NM_021072.4, c.1195T>C, p.(Ser399Pro). This variant might have a dominant-negative effect on channel function, leading to severe epileptic
encephalopathy. SIMILAR CONTENT BEING VIEWED BY OTHERS DE NOVO Y1460C MISSENSE VARIANT IN NAV1.1 IMPEDES THE PORE REGION AND RESULTS IN EPILEPTIC ENCEPHALOPATHY Article Open access 13
October 2022 SCN8A EPILEPTIC ENCEPHALOPATHY MUTATIONS DISPLAY A GAIN-OF-FUNCTION PHENOTYPE AND DIVERGENT SENSITIVITY TO ANTIEPILEPTIC DRUGS Article 27 July 2022 A NOVEL DE NOVO _KCNB1_
VARIANT ALTERING CHANNEL CHARACTERISTICS IN A PATIENT WITH PERIVENTRICULAR HETEROTOPIA, ABNORMAL CORPUS CALLOSUM, AND MILD SEIZURE OUTCOME Article 18 October 2022 Hyperpolarization-activated
cyclic nucleotide-gated (HCN) channels mediate a cationic current that stabilizes the neuronal membrane potential against excitatory or inhibitory input and regulates neuronal network
excitability1,2. _HCN1_ (NM_021072), one of the four genes that encode HCN channels1, is highly expressed in the neocortex, hippocampus, and brainstem of the central nervous system3. It was
therefore presumed that pathogenic _HCN1_ variants could produce pharmacoresponsive epilepsy or developmental and epileptic encephalopathy (DEE). In 2014, Nava _et al_. identified five
different _de novo HCN1_ variants in patients with Dravet-like syndrome without _SCN1A_ and _PCDH19_ variants or fever-sensitive epileptic encephalopathy4. Additionally, recent studies have
revealed that the phenotypic spectrum associated with _HCN1_ variants ranges from genetic epilepsy with febrile seizures plus (OMIM #618482) or genetic generalized epilepsy to neonatal- or
infantile-onset DEE5,6,7,8,9,10 (OMIM #615871). Herein, we report a Japanese patient with a de novo heterozygous _HCN1_ variant who had presented since early infancy with repetitive focal
seizures and recurrent super-refractory status epilepticus, which were associated with profound developmental delay. This report provides detailed clinical findings that expand the known
phenotypic spectrum of _HCN1_-related DEE. The male patient, the second child of unrelated parents, was born at 39 weeks gestational age after a normal pregnancy and delivery. His birth
weight, length, and occipitofrontal circumference were 3380 g (+0.8 standard deviation score [SDS]), 50.8 cm (+0.9 SDS), and 33.8 cm (+0.3 SDS), respectively. His father, elder brother, and
maternal uncle had a history of febrile seizures. At 2 months of age, the patient developed convulsive seizures that gradually increased in frequency, occurring in clusters despite
administration of phenobarbital and sodium valproate. He was first admitted to our hospital at 4 months of age. His anthropometric measurements were as follows: body weight 7.6 kg (+0.6
SDS), height 60.5 cm (−1.6 SDS), and occipitofrontal circumference 42.4 cm (+1.3 SDS). Psychomotor development was normal. After admission, right or left hemi-clonic seizures and focal to
bilateral tonic–clonic seizures were observed. Shortly afterward, a respiratory infection elicited a high fever and provoked the onset of convulsive status epilepticus. Intravenous
anti-seizure medications, including diazepam, midazolam, phenytoin, and lidocaine, were ineffective. Ultimately, thiopental was effective in terminating seizure activity. An interictal
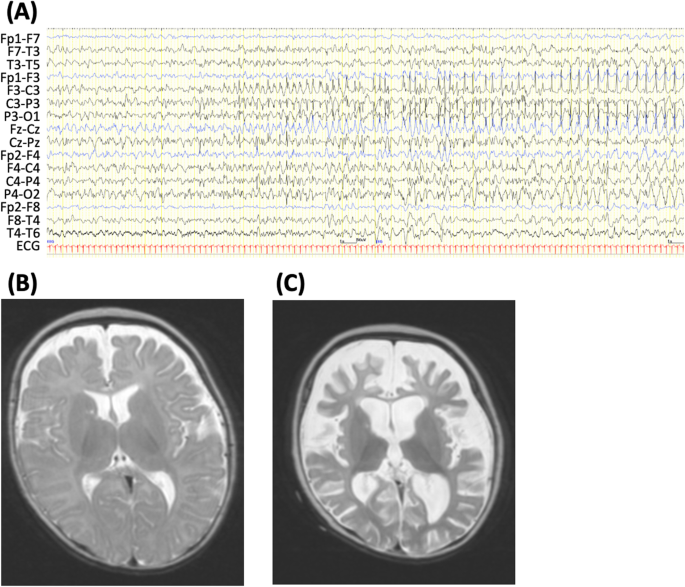
electroencephalogram (EEG) showed spike discharges in the left frontal area. An ictal EEG revealed spike-wave discharge with onset in the left fronto-centro-parietal region (Fig. 1A) or
right fronto-central region. Laboratory examinations and brain magnetic resonance imaging (MRI) findings showed no abnormalities at that time (Fig. 1B). At 6 months of age, during another
respiratory tract infection, the patient developed a prolonged generalized tonic convulsion lasting 30 min, which evolved into repetitive generalized convulsions with prolonged hypoxia. This
prolonged seizure was refractory to anticonvulsants, including thiopental, and lasted 36 h. Pentobarbital was given intravenously, and mechanical ventilation was required for 2 weeks. After
extubation, seizures were controlled by high-dose phenobarbital treatment combined with potassium bromide and zonisamide. However, the patient’s motor and cognitive development were
severely impaired. Brain MRI at 8 months of age showed severe cortical and white matter atrophy (largely in the frontal lobe) and ventricular enlargement (Fig. 1C). At 10 months of age,
brief daily seizures involving grinning or a pale face were noted. At 13 years of age, the patient had generalized hypotonia and spastic quadriplegia with profound intellectual and motor
impairment. He could not control his head, spoke no meaningful words and was almost entirely bedridden. An increase in brief seizures with tonic movements elicited the addition of
levetiracetam and topiramate to existing medications (phenobarbital, potassium bromide, and zonisamide). However, these medications were only partially effective. At 18 years of age, the
patient continued to have brief focal seizures, often in clusters requiring intravenous phenobarbital for seizure control. Whole exome sequencing was performed using a SureSelectXT Human All
Exon v5 (Agilent Technologies, Santa Clara, CA), and captured libraries were sequenced using an Illumina HiSeq 2500 (Illumina, San Diego, CA) with 101 base-paired end reads. Exome data
processing, variant calling, and variant annotation were performed as previously described11. Variant pathogenicity was predicted using SIFT, Polyphen-2, CADD and M-CAP (Table S1). Whole
exome sequencing of the patient’s leukocyte-derived genomic DNA identified an _HCN1_ variant: NM_021072.4, c.1195T>C, p.(Ser399Pro). Trio-based Sanger sequencing confirmed that this was a
de novo variant. The variant was absent in 38 K JPN and gnomAD v2.1.1 (accessed on 22nd Feb 2023), as well as in our 408 in-house control exomes (all Japanese). The same variant was
previously reported in a patient with infantile epileptic encephalopathy who had prolonged febrile seizures and intractable apneic tonic–clonic seizures starting at 4 months of age5.
Therefore, the variant was classified as pathogenic according to the ACMG-AMP Guidelines (PS1, PS2, PM2, and PP3). The clinical and molecular genetic studies were performed in accordance
with the Declaration of Helsinki and were approved by the institutional review board of Yamagata University Faculty of Medicine, Showa University School of Medicine, and Yokohama City
University School of Medicine. The patient’s parents provided written informed consent. We compared the features of the present patient with those of a previous patient carrying the same
genetic variant5 and summarized the clinical manifestations of previously reported cases10 (Table 1). The present patient showed infantile-onset DEE with repetitive focal seizures, recurrent
refractory status epilepticus, and progressive brain atrophy. Among previously reported patients with _HCN1_ variants, brain atrophy was observed in only one patient with a p.G391D
variant5,10. Therefore, the phenotype in the present patient appears to be more severe than that of a previous patient with the same p.S399P variant. This phenotypic variability may be
caused by genetic modifiers such as a prominent family history of febrile seizures even though no other variants in the known epilepsy-related genes were identified through whole exome
sequencing of this patient. Progressive brain atrophy may also have been caused by acute encephalopathy or encephalitis. However, because MRI changes were not observed during the acute phase
of status epilepticus in the patient, it was unlikely that he had acute encephalopathy or encephalitis. Recurrent status epilepticus, refractory to intravenous anticonvulsant
administration, was one of the characteristic features observed in this patient. Administration of general anesthesia was necessary to abolish a prolonged seizure lasting 36 hours. In
previous studies, status epilepticus was reported in 5/18 patients with loss-of-function _HCN1_ variants and in 1/13 patients with gain-of-function _HCN1_ variants9. The p.S399P variant is
also a loss-of-function variant. However, status epilepticus is caused not by a specific _HCN1_ variant but by a variety of variants9. Both upregulation and downregulation of HCN channels
have been associated with epileptic activity in animal models5,12. A previous study indicated that patients with the p.G391D variant showed the most severe phenotypes among _HCN1_ DEE5
patients, similar to the present patient with the p.S399P variant. HCN subunits have six transmembrane domains (S1–S6), including a positively charged voltage sensor (S4) and the
ion-conducting pore region between S5 and S63. The amino acid glycine 391, located on the intracellular interface of the S6 transmembrane domain, is adjacent to the p.S399P variant5.
Transient expression of _HCN1_ variants in Chinese hamster ovary cells showed that the protein levels of both the p.G391D and p.S399P variants were significantly decreased compared with
those of wild-type proteins. In addition, whole-cell patch-clamp recordings showed no current in cells separately transfected with each variant, suggesting that these are loss-of-function
variants5. Interestingly, when cells were cotransfected with wild-type and p.G391D variant constructs, a strong reduction in current density was observed, suggesting that p.G391D has a
dominant-negative effect on heteromeric channel function. Similarly, severe DEE in the present patient may have been caused by a dominant-negative effect of the p.S399P variant. The
epileptic seizure frequency in patients with _HCN1_ variants ranges from no to daily seizures, and half of patients demonstrate resistance to anti-seizure medications5. High-dose
phenobarbital and potassium bromide were partially effective for the present patient. In murine models of p.G391D and p.M153I, administration of the sodium channel antagonists lamotrigine
and phenytoin resulted in paradoxical induction of seizures; however, administration of sodium valproate did not lead to convulsive seizures13. In _Hcn1_M294L mice carrying a homolog of the
_HCN1_ p.M305L variant; phenytoin, lamotrigine, retigabine, and carbamazepine increased spike frequency, whereas levetiracetam, diazepam, sodium valproate, and ethosuximide significantly
reduced spike frequency14. Sodium channel blockers such as carbamazepine or phenytoin may aggravate seizures in Dravet syndrome with _SCN1A_ variants15. These blockers might also aggravate
seizures in patients with _HCN1_ variants; therefore, early genetic diagnosis could be crucial for appropriate pharmacotherapy selection. HGV DATABASE The relevant data from this Data Report
are hosted at the Human Genome Variation Database at https://doi.org/10.6084/m9.figshare.hgv.3311. REFERENCES * Biel, M., Wahl-Schott, C., Michalakis, S. & Zong, X.
Hyperpolarization-activated cation channels: from genes to function. _Physiol. Rev._ 89, 847–885 (2009). Article CAS PubMed Google Scholar * Poolos, N. P. Hyperpolarization-activated
cyclic nucleotide-gated (HCN) ion channelopathy in epilepsy. In: Noebels, J. L., Avoli, M., Rogawski, M. A., Olsen, R. W., & Delgado-Escueta, A. V. (eds). _Jasper’s Basic Mechanisms of
the Epilepsies_, 4th edn. Oxford University Press: Oxford, USA, 85–96 (2012). * Benarroch, E. E. HCN channels: function and clinical implications. _Neurology_ 80, 304–310 (2013). Article
PubMed Google Scholar * Nava, C. et al. De novo mutations in _HCN1_ cause early infantile epileptic encephalopathy. _Nat. Genet._ 46, 640–645 (2014). Article CAS PubMed Google Scholar
* Marini, C. et al. _HCN1_ mutation spectrum: from neonatal epileptic encephalopathy to benign generalized epilepsy and beyond. _Brain_ 141, 3160–3178 (2018). Article PubMed Google Scholar
* Parrini, E. et al. Diagnostic targeted resequencing in 349 Patients with drug-resistant pediatric epilepsies identifies causative mutations in 30 different genes. _Hum. Mutat._ 38,
216–225 (2017). Article CAS PubMed Google Scholar * Bonzanni, M. et al. A novel _de novo HCN1_ loss-of-function mutation in genetic generalized epilepsy causing increased neuronal
excitability. _Neurobiol. Dis._ 118, 55–63 (2018). Article CAS PubMed Google Scholar * Porro, A. et al. Do the functional properties of _HCN1_ mutants correlate with the clinical
features in epileptic patients? _Prog. Biophys. Mol. Biol._ 166, 147–155 (2021). Article CAS PubMed Google Scholar * Xie, C. et al. Novel _HCN1_ mutations associated with epilepsy and
impacts on neuronal excitability. _Front. Mol. Neurosci._ 15, 870182 (2022). Article CAS PubMed PubMed Central Google Scholar * Kessi, M. et al. The contribution of HCN channelopathies
in different epileptic syndromes, mechanisms, modulators, and potential treatment targets: a systematic review. _Front. Mol. Neurosci._ 15, 807202 (2022). Article CAS PubMed PubMed
Central Google Scholar * Saitsu, H. et al. De novo mutations in the autophagy gene _WDR45_ cause static encephalopathy of childhood with neurodegeneration in adulthood. _Nat. Genet._ 45,
445–449.e1 (2013). Article CAS PubMed Google Scholar * Noam, Y., Bernard, C. & Baram, T. Z. Towards are integrated view of HCN channel role in epilepsy. _Curr. Opin. Neurobiol._ 21,
873–879 (2011). Article CAS PubMed PubMed Central Google Scholar * Merseburg, A. et al. Seizures, behavioral deficits, and adverse drug responses in two new genetic mouse models of
_HCN1_ epileptic encephalopathy. _eLife_ 11, e70826 (2022). Article CAS PubMed PubMed Central Google Scholar * Bleakley, L. E., McKenzie, C. E. & Reid, C. A. Efficacy of antiseizure
medication in a mouse model of _HCN1_ developmental and epileptic encephalopathy. _Epilepsia_ 64, e1–e8 (2023). Article CAS PubMed Google Scholar * Wirrell, E. C. et al. International
consensus on diagnosis and management of Dravet syndrome. _Epilepsia_ 63, 1761–1777 (2022). Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS We
thank the patient and his family for their participation in this study. This work was supported by the Japan Agency for Medical Research and Development (AMED) (grant numbers JP22ek0109486,
JP22ek0109549, JP22ek0109493) and the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant-in-Aid for Scientific Research (B) (grant number JP20H03641) and (C) (grant number
JH21K06819). We thank Edanz (https://jp.edanz.com/ac) for editing the English text of a draft of this manuscript. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Child Neurology,
National Hospital Organization Nishiniigata Chuo Hospital, Niigata, Japan Yu Kobayashi, Jun Tohyama, Kei Yamada, Moemi Hojo, Eijun Seki, Masaki Miura & Noriko Soma * Department of
Pediatrics, Niigata Prefecture Hamagumi Medical Rehabilitation Center for Disabled Children, Niigata, Japan Noriyuki Akasaka * Department of Pediatrics, Niigata University Medical and Dental
Hospital, Niigata, Japan Takeshi Ono * Department of Pediatrics, Showa University School of Medicine, Tokyo, Japan Mitsuhiro Kato * Department of Biochemistry, Hamamatsu University School
of Medicine, Hamamatsu, Japan Mitsuko Nakashima & Hirotomo Saitsu * Department of Human Genetics, Yokohama City University Graduate School of Medicine, Yokohama, Japan Naomichi Matsumoto
Authors * Yu Kobayashi View author publications You can also search for this author inPubMed Google Scholar * Jun Tohyama View author publications You can also search for this author
inPubMed Google Scholar * Noriyuki Akasaka View author publications You can also search for this author inPubMed Google Scholar * Kei Yamada View author publications You can also search for
this author inPubMed Google Scholar * Moemi Hojo View author publications You can also search for this author inPubMed Google Scholar * Eijun Seki View author publications You can also
search for this author inPubMed Google Scholar * Masaki Miura View author publications You can also search for this author inPubMed Google Scholar * Noriko Soma View author publications You
can also search for this author inPubMed Google Scholar * Takeshi Ono View author publications You can also search for this author inPubMed Google Scholar * Mitsuhiro Kato View author
publications You can also search for this author inPubMed Google Scholar * Mitsuko Nakashima View author publications You can also search for this author inPubMed Google Scholar * Hirotomo
Saitsu View author publications You can also search for this author inPubMed Google Scholar * Naomichi Matsumoto View author publications You can also search for this author inPubMed Google
Scholar CONTRIBUTIONS Y.K. cared for the patient and drafted the manuscript. N.A., K.Y., M.H., E.S., N.S., and T.O. cared for the patient. M.K. helped draft the manuscript and critically
revised the manuscript for important content. M.N., H.S., and N.M. were responsible for genomic analysis, critically revised the manuscript for important content, and helped draft the
manuscript. J.T. cared for the patient and revised the manuscript. All authors contributed to the writing of the final manuscript. CORRESPONDING AUTHOR Correspondence to Jun Tohyama. ETHICS
DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in
published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTAL TABLE 1. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons
Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original
author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the
article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use
is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Kobayashi, Y., Tohyama, J., Akasaka, N. _et al._ The _HCN1_ p.Ser399Pro variant
causes epileptic encephalopathy with super-refractory status epilepticus. _Hum Genome Var_ 10, 20 (2023). https://doi.org/10.1038/s41439-023-00247-8 Download citation * Received: 24 April
2023 * Revised: 05 June 2023 * Accepted: 05 June 2023 * Published: 23 June 2023 * DOI: https://doi.org/10.1038/s41439-023-00247-8 SHARE THIS ARTICLE Anyone you share the following link with
will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt
content-sharing initiative