Play all audios:
ABSTRACT Variants predicted to result in the loss of function of human genes have attracted interest because of their clinical impact and surprising prevalence in healthy individuals. Here,
we present ALoFT (annotation of loss-of-function transcripts), a method to annotate and predict the disease-causing potential of loss-of-function variants. Using data from Mendelian
disease-gene discovery projects, we show that ALoFT can distinguish between loss-of-function variants that are deleterious as heterozygotes and those causing disease only in the homozygous
state. Investigation of variants discovered in healthy populations suggests that each individual carries at least two heterozygous premature stop alleles that could potentially lead to
disease if present as homozygotes. When applied to de novo putative loss-of-function variants in autism-affected families, ALoFT distinguishes between deleterious variants in patients and
benign variants in unaffected siblings. Finally, analysis of somatic variants in >6500 cancer exomes shows that putative loss-of-function variants predicted to be deleterious by ALoFT are
enriched in known driver genes. SIMILAR CONTENT BEING VIEWED BY OTHERS A DEEP CATALOGUE OF PROTEIN-CODING VARIATION IN 983,578 INDIVIDUALS Article Open access 20 May 2024 ASSESSMENT OF
ABILITY OF ALPHAMISSENSE TO IDENTIFY VARIANTS AFFECTING SUSCEPTIBILITY TO COMMON DISEASE Article Open access 03 August 2024 LOSS-OF-FUNCTION, GAIN-OF-FUNCTION AND DOMINANT-NEGATIVE MUTATIONS
HAVE PROFOUNDLY DIFFERENT EFFECTS ON PROTEIN STRUCTURE Article Open access 06 July 2022 INTRODUCTION One of the most notable findings from personal genomics studies is that all individuals
harbor loss-of-function (LoF) variants in some of their genes1. A systematic study of LoF variants from the 1000 Genomes Project revealed that there are over 100 putative LoF (pLoF) variants
in each individual2,3,4. Recently, a larger study aimed at elucidating rare LoF events in 2636 Icelanders generated a catalog of 1171 genes that contain either homozygous or compound
heterozygous LoF variants with a minor allele frequency less than 2%5. Thus, several genes are knocked out either completely or in an isoform-specific manner. The discovery of protective LoF
variants associated with beneficial traits and their potential to enable identification of valuable drug targets has fueled an increased interest in pLoF variants. For example, nonsense
variants in _PCSK9_ are associated with low low-density lipoprotein (LDL) levels6, which prompted the active pursuit of the inhibition of _PCSK9_ as a potential therapeutic for
hypercholesterolemia7, 8 and led to the development of two drugs that have been recently approved by the FDA. Other examples include nonsense and splice mutations in _APOC3_ associated with
low levels of circulating triglycerides, a nonsense mutation in _SLC30A8_ resulting in about 65% reduction in risk for Type II diabetes, two splice variants in the Finnish population in
_LPA_ that protect against coronary artery disease, and two LoF-producing splice variants and a nonsense mutation in _HAL_ associated with increased blood histidine levels and reduced risk
of coronary artery disease9,10,11,12. About 12% of known disease-causing mutations in the Human Gene Mutation Database (HGMD) are due to nonsense mutations13. Even though premature stop
variants often lead to loss of function and are thus deleterious, predicting the functional impact of premature stop codons is not straightforward. Aberrant transcripts containing premature
stop codons are typically removed by nonsense-mediated decay (NMD), an mRNA surveillance mechanism14. However, a recent large-scale expression analysis demonstrated that 68% of predicted NMD
events due to premature stop variants are unsupported by RNA-Seq analyses15. Moreover, premature stop codons in the last exon are generally not subject to NMD. A study aimed at
understanding disease mutations using a 3D structure-based interaction network suggests that truncating mutations can give rise to functional protein products16. Furthermore, when a variant
affects only some isoforms of a gene, it is difficult to infer its impact on gene function without the knowledge of the isoforms that are expressed in the tissue of interest and how their
levels of expression affect gene function. Finally, loss of function of a gene might not have any impact on the fitness of the organism. While there are several algorithms to predict the
effect of missense coding variants on protein function, there is a paucity of methods that are applicable to nonsense variants17,18,19. Additionally, current prediction methods that infer
the pathogenicity of variants do not take into account the zygosity of the variant20, 21. The majority of pLoF variants in healthy cohorts are heterozygous. It is likely that a subset of
these variants will cause disease as homozygotes. Here we present a pipeline called ALoFT (Annotation of Loss-of-Function Transcripts), that provides extensive annotation of pLoF variants.
Furthermore, we developed a prediction model to classify pLoF variants into three classes: those that are benign, those that lead to recessive disease (disease-causing only when homozygous)
and those that lead to dominant disease (disease-causing as heterozygotes). Finally, we validated the prediction model by applying ALoFT to known disease mutations in Mendelian diseases,
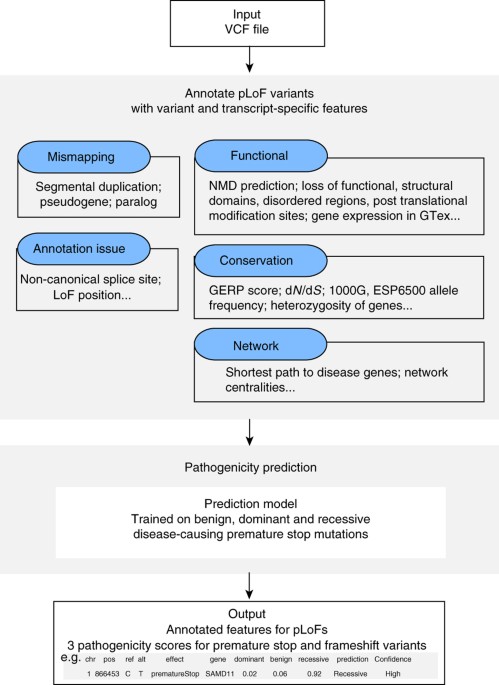
autism, and cancer. RESULTS ALOFT PIPELINE We have developed a pipeline called ALoFT to annotate pLoF variants. In this study, we included premature stop-causing single-nucleotide
polymorphisms (SNPs), frameshift-causing indels and variants affecting canonical splice sites as pLoF variants, also referred to as protein truncating variants. An overview of the pipeline
is shown in Supplementary Fig. 1. The main features of ALoFT include (1) functional domain annotations; (2) evolutionary conservation; and (3) biological networks. For comprehensive
functional annotation, we integrated several annotation resources such as PFAM and SMART functional domains22, 23, signal peptide and transmembrane annotations, post-translational
modification sites, NMD prediction24, 25, and structure-based features such as SCOP domains and disordered residues. For evolutionary conservation, ALoFT outputs variant position-specific
GERP scores, which is a measure of evolutionary conservation26 and d_N_/d_S_ values (ratio of missense to synonymous substitution rates) for macaque and mouse that are computed from
human-macaque and human-mouse orthologous alignments, respectively. In addition, we evaluated if the region removed due to the truncation of the coding sequence is evolutionarily conserved
based on constrained elements27. ALoFT includes network features shown to be important in disease prediction algorithms: a proximity parameter that gives the number of disease genes
connected to a gene in a protein–protein interaction network and the shortest path to the nearest disease gene2, 28. The pipeline also includes features to help identify erroneous LoF calls,
potential mismapping, and annotation errors, because LoF variant calls have been shown to be enriched for annotation and sequencing artifacts2. A description of all the annotations provided
by ALoFT is included in Supplementary Table 1 (details in Methods). Documentation and link to source code can be found at aloft.gersteinlab.org. Using the annotations output by ALoFT as
predictive features (Fig. 1, Supplementary Data 1), we developed a prediction method to infer the pathogenicity of pLoF variants. To build the ALoFT classifier, we used three classes of
premature stop variants as training data: benign variants, dominant disease-causing variants, and recessive disease-causing variants (Supplementary Table 2). The benign set includes
homozygous premature stop variants discovered in a cohort of 1092 healthy people, Phase1 1000 Genomes data (1KG). Homozygous premature stop mutations from HGMD that lead to recessive disease
and heterozygous premature stop variants in haplo-insufficient genes that lead to dominant disease represent the two disease classes3, 28. In addition to loss-of-function effects,
truncating mutations can also lead to gain of function. However, gain-of-function mutations are difficult to model systematically as the effect of a variant can only be understood in the
context of the biology of the gene and can vary widely for different genes and gene classes. In order to minimize errors that might arise due to inadequate modeling of gain-of-function
effects and to focus on LoF, we only use predicted haploinsufficient genes as the training data for the dominant model. We built the ALoFT classifier to distinguish among the three classes
using a random forest algorithm29 (details in Methods). For each mutation, ALoFT provides three class probability estimates, and we obtain good discrimination between each class. The
prediction output provides the three scores for each pLoF variant that correspond to the probability of the pLoF being benign, dominant or recessive disease-causing allele. In addition,
ALoFT also provides the predicted pathogenicity. The pathogenic effect of pLoF variant is assigned to the class that corresponds to the maximum score. EVALUATION OF THE CLASSIFIER The
average multiclass test area under the curve (AUC) with 10-fold cross-validation is 0.97. The precision for the three classes are as follows: dominant = 0.86, recessive = 0.86, and benign =
0.96. The recall for the three classes are as follows: dominant: 0.71; recessive: 0.95; and benign: 0.96. The classifier is robust to the choice of training data sets (Supplementary Table 3,
details in Methods). Though trained with premature stop SNVs, our method is also applicable to frameshift indels. We applied ALoFT to classify pathogenic indels in HGMD. 99.4% of HGMD
disease-causing frameshift indels are predicted to be pathogenic based on the maximum ALoFT score. We analyzed the importance of the various features to the classification (Supplementary
Fig. 2). The global allele frequency of variants in the Exome Aggregation Consortium, ExAC, a data set comprising sequence variations obtained from an analysis of 60,706 unrelated
individuals of diverse ethnicities (ExAC30, http://exac.broadinstitute.org), appears to be the most important feature for classification. When we removed this feature and other features
related to allele frequency (i.e., features related to variants in both ExAC and Exome Sequencing Project, ESP) and retrained the random forest model, the classifier still performs well with
an average multiclass test AUC of 0.93. (The precisions for the three classes are as follows: dominant = 0.84, recessive = 0.80, and benign = 0.75). We also systematically evaluated the
classifier using models trained on varying sets of features (Supplementary Table 4). Overall, we find that the classification is not driven by any single feature and integrating many
features improves prediction accuracy. VALIDATION OF THE CLASSIFIER We applied ALoFT to elucidate the pathogenicity of pLoF variants in various disease scenarios. Using case studies, we show
that ALoFT provides robust predictions for the effect of pLoFs. UNDERSTANDING PLOFS IN MENDELIAN DISEASE We evaluated ALoFT by predicting the effect of known disease-causing premature stop
mutations from ClinVar31 (details in Methods) and predicted the mode of inheritance and pathogenicity of all of truncating variants (Fig. 2a). ALoFT is clearly able to distinguish between
pLoFs that lead to disease in a heterozygous state vs. those that do so only in a homozygous state. Our method shows that heterozygous disease-causing variants have significantly higher
dominant disease-causing scores than the homozygous disease-causing variants (_p_-value: 1.3e-13; Wilcoxon rank-sum test). We used two other measures, GERP score, which is a measure of
evolutionary conservation, and CADD score, which gives a measure of pathogenicity, to classify recessive vs. dominant pLoF variants32. Both CADD (_p_-value: 0.13; Wilcoxon rank-sum test) and
GERP (_p_-value: 0.49; Wilcoxon rank-sum test) scores are not able to discriminate between recessive and dominant disease-causing mutations (Fig. 2a). We also tested our method on a smaller
data set from the Center For Mendelian Genomics studies33 and were able to correctly recapitulate the pathogenic effect of pLoF variants and their inheritance pattern (Fig. 2b).
UNDERSTANDING DE NOVO PLOFS IMPLICATED IN AUTISM De novo pLoF SNPs have been implicated in autism based on analysis of sporadic or simplex families (families with no prior history of
autism)34,35,36,37. We applied our method to de novo pLoF mutations discovered in these studies. Each individual carries about one de novo premature stop variant (Supplementary Table 5). Our
method shows that the proportion of dominant disease-causing de novo LoF events is significantly higher in autism patients vs. siblings of patients with autism (Fig. 3a; _p_-value: 8.4e−4;
Wilcoxon rank-sum test). Autism spectrum disorder is known to be four times more prevalent in males than in females suggesting a protective effect in females. Previous studies show that a
higher mutational burden of non-synonymous mutations is ascertained in females with autism spectrum disorder38. Therefore, we investigated differences in the impact of de novo pLoF variants
in male vs. female autism patients. We observed a similar pattern for pLoF mutations as has been found for missense variants—female probands have a higher proportion of predicted deleterious
de novo pLoF variants than male probands (Fig. 3a; _p_-value: 0.03; Wilcoxon rank-sum test). Supplementary Data 2 includes the ALoFT predictions for de novo pLoF variants. A recent study
based on exome sequencing of 3871 autism cases delineated 33 risk genes at FDR < 0.139. We observed that de novo pLoF mutations in the 33 risk genes of the autism patients have higher
dominant disease-causing scores than the de novo pLoF variants in other genes (Fig. 3b; _p_-value: 5e−3; Wilcoxon rank-sum test). Thus, ALoFT predictions corroborate the role of de novo pLoF
variants in autism as shown by others using entirely different approaches. IDENTIFICATION OF PATHOGENIC SOMATIC LOF VARIANTS IN CANCER We applied our prediction method to infer the effect
of somatic premature stop variants (somatic pLoFs) from a compilation of 6535 cancer exomes40. As shown in Fig. 4, somatic pLoFs are enriched in known cancer driver genes compared to
randomly sampled genes of matched lengths. Moreover, deleterious somatic LoFs are strongly enriched in driver genes and depleted in LoF-tolerant genes (genes that contain at least one
homozygous LoF variant in the 1KG population). In the context of somatic mutations, variant zygosity, or distinguishing between ‘dominant’ and ‘recessive’ disease-causing mutations, is not
always relevant. Cancer cells may show aneuploidy and cellular heterogeneity. Therefore, for the evaluation of somatic mutations, we define an overall measure of deleteriousness as (1-benign
ALoFT score) on the _X_ axis of Fig. 4. We also evaluated ALoFT as a tool for distinguishing driver LoF mutations from passenger LoF mutations in tumors with high mutation burden. We
observed a decrease in deleterious LoF mutations with increasing total mutational burden (Fig. 5a). However, the ratio of deleterious LoFs to total pLoFs displayed no significant trend
across groups (Fig. 5b). The ratio of deleterious LoF mutations to total pLoF mutations is consistently high across groups (84%). This may indicate that driver LoF events tend to arise early
in tumor development. To classify genes as tumor suppressors, Vogelstein et al.41 proposed a “20/20” rule, whereby a gene is classified as a tumor suppressor if >20% of the observed
mutations in that gene are inactivating mutations. Among the 210 genes that met 20/20 rule criteria, 87% of pLoF mutations affecting these genes were deleterious LoFs, representing 21% of
total mutations. By comparison, only 6% of mutations were deleterious LoFs among 11,892 genes that did not meet 20/20 criteria (_p_ < 0.001, chi-squared test) (Fig. 6). A list of these
genes is provided as Supplementary Data 3. In cases where genes display a high somatic pLoF rate but low somatic deleterious LoF rate, ALoFT may be used to identify potential false-positive
driver genes predicted by the 20/20 rule. DISTINGUISHING BETWEEN BENIGN AND PATHOGENIC PLOFS Finally, we applied ALoFT to predict the effect of premature stop variants in the final exons of
protein-coding genes. It is often assumed that premature stop variants in the last coding exon are likely to be benign because they could escape NMD; as a result, in many cases, the effect
will be the expression of a truncated protein rather than a complete loss of function. However, several examples of disease-causing mutations in the last exon are known42. Therefore, we
applied ALoFT to see if we could distinguish between benign and disease-causing LoF variants in the last coding exon. To this end, we applied ALoFT to understand the effect of pLoF variants
in ESP6500, ExAC, and HGMD data sets. A higher proportion of rare variants is observed in ESP6500 and ExAC cohorts due to their larger sample size and higher sequencing depth (Fig. 7a). A
large number of both common and rare premature stop variants are seen at the end of the coding genes in the 1KG, ESP6500, and ExAC data sets. In contrast, fewer disease-causing HGMD variants
are seen at the ends of coding genes (Fig. 7b). ALoFT predicts that both common and rare premature stop variants in the last coding exon in the 1KG, ESP6500, and ExAC cohort are likely to
be benign, whereas HGMD mutations in the last coding exon tend to be disease-causing (Fig. 7b). Thus, ALoFT is able to differentiate between rare benign premature stop variants seen in
healthy individuals and rare disease-causing HGMD alleles. PLOFS IN AN INDIVIDUAL GENOME The above case studies clearly illustrate the validity of the ALoFT score in elucidating the effect
of pLoF variants. In order to estimate the number of pLoF disease alleles in a healthy individual, we applied ALoFT to premature stop variants from the 1KG and ExAC data sets. The predicted
benign score for pLoFs in 1KG has a wide range of values (Fig. 8, Supplementary Data 4). Furthermore, due to differences in sequencing coverage and variant calling approaches, the number of
potential disease pLoFs per individual varies among datasets. In general, the number increases with higher coverage and larger cohorts where joint variant calling methods result in improved
sensitivity in the identification of rare variants. To conservatively estimate a lower bound for per individual statistics (Methods), we applied a stringent filtering strategy to restrict to
high confidence pLoFs. On average, each individual is a carrier of at least two rare heterozygous premature stop alleles that are predicted to be disease-causing in the homozygous state
(Supplementary Table 6) based on the 1KG Phase1 data. Current estimates of the genetic burden of disease alleles (all types of variation, including LoFs) in an individual vary widely,
ranging from 1.1 recessive alleles per individual to 31 deleterious alleles43,44,45,46,47. In connection with this, it should be noted that the referenced studies are based on diverse
methods of identifying variants ranging from targeted panel-based candidate gene studies to whole-genome sequencing. The estimation of the number of deleterious pLoF alleles can be affected
by a number of confounding factors that include incomplete penetrance of disease alleles, variable expressivity, compensatory mutations, marginal variant calls, and imperfect training data
sets (Methods). Next, we looked at premature stop variants in the 1KG cohort in known disease-causing genes. We find that variants in 1KG are more likely to be benign compared to known
disease-causing mutations in the same genes (Fig. 8; green vs. blue boxes, _p_-value: 6.9e−9). Our results provide a possible rationale for this observation. Firstly, variants predicted to
be benign in 1KG often affect isoforms that are different from the isoforms containing the disease-causing HGMD variant. This suggests that LoFs in healthy individuals may affect minor
isoforms (Supplementary Fig. 3). About 12.4% of premature stop variants in the presumed healthy 1KG individuals occur in known disease genes and the disease-causing variants in the same
genes are on different isoforms. Secondly, some variants predicted to be benign in 1KG occur in the last exon or later in the protein-coding transcript relative to the disease-causing
variant in the same transcript. The effect of such variants is possibly the production of truncated proteins that are sufficiently functional. Lastly, a majority of 1KG variants seen in
disease genes are predicted to be disease-causing only if they are homozygous. However, they occur as rare heterozygous variants in the 1KG cohort. Mutations in HGMD are assumed to be
disease-causing. However, some mutations are predicted to be benign by ALoFT (Fig. 8). It is known that disease databases include incorrect disease annotations and common variants and about
27% of variants were excluded by Bell et al.43 in their estimate of carrier burden for severe recessive diseases. However, overall only 0.67% of HGMD premature stop mutations are predicted
to be benign. Supplementary Fig. 4 shows that most mutations predicted to be benign by ALoFT are seen at higher allele frequencies than those predicted to be in the dominant and recessive
classes. Of the 119 pLoF autosomal variants in HGMD predicted to be benign by ALoFT, 32 variants are in Filaggrin, _FLG_. _FLG_ LoF mutations are linked to susceptibility to atopic
dermatitis, a skin condition leading to eczema. Eczema is a complex trait and the resulting phenotypes are highly variable due to the interplay of environmental and genetic factors. A recent
study showed that individuals with bi-alleleic null variants of _FLG_ do not always have atopic dermatitis48. A study on British Pakistanis with related parents identified 781 genes
containing rare homozygous LoF variants49. They found homozygous LoF variants in recessive Mendelian disease genes; however, carriers of most of these homozygous LoF variants do not have the
disease phenotype. We applied ALoFT to classify these homozygous LoF variants. Of the 22 variants for which ALoFT provides predictions, 3 are predicted to be benign and none of them were
predicted to lead to disease by the dominant mode of inheritance. However, 19 homozygous variants are indeed predicted to lead to disease with a recessive mode of inheritance (Supplementary
Data 5). The lack of a discernible phenotype could be due to incomplete penetrance of the mutations or due to modifier effects. The penetrance of some disease mutations are also known to be
age dependent and sex dependent50. While studies in consanguineous populations have been used to identify recessive disease genes51, the absence of disease provides an opportunity to look
for modifiers in their genetic background. DISCUSSION In summary, we describe ALoFT, a tool for predicting the impact of pLoF variants. In the context of a diploid model, it may be used to
determine whether pLoF variants are likely to lead to recessive or dominant disease. Better identification and characterization of pLoF variants have both diagnostic and therapeutic
implications. ALoFT allows for the identification and prioritization of high-impact putative disease-causing pLoF variants in individual genomes. Integrating benign LoF variants with
phenotypic information will help us to identify protective variants that are valuable drug targets52. Gene functions important for species propagation might actually be deleterious as one
ages; thus, LoF variants in such genes provide an intriguing avenue to discover targets for aging-related diseases53. Lastly, diseases caused by LoF variants provide opportunities for
targeted therapy using drugs that either enable read-through of the premature stop, thus restoring the function of the mutant protein, or NMD inhibitors that prevent degradation of the
LoF-containing transcript by NMD54, 55. This is especially useful in the context of rare diseases where targeting the same molecular phenotype leading to different diseases alleviates the
need to design a new drug for each individual disease. Further work will be needed both to correlate the predictions of ALoFT with experimental assays of protein LoF and to study the
phenotypic impact of heterozygous and homozygous LoF variants in large clinical cohorts. METHODS OVERVIEW OF ALOFT ANNOTATION PIPELINE ALoFT provides extensive annotation for SNPs that
introduce a premature stop codon, SNPs affecting splice sites, and indels that lead to frameshift. Initial sequence-based annotation of coding variants is performed by the Variant Annotation
Tool56 (VAT). The output of VAT is augmented with various features specific to pLoF variants. The input files can be in VCF format or a tab-delimited 5-column file that includes chromosome,
variant position, variant ID, reference allele, and alternate allele. LoF variants annotated with various features are output as three separate files: a VCF-formatted file containing
summarized annotations, a Tab-delimited file containing extensive annotations for premature stop variants and indels leading to frameshift, a tab-delimited file containing annotations for
variants that affect the canonical splice sites. The output of ALoFT annotation pipeline is discussed below and the overview of the pipeline is shown in Supplementary Fig. 1. FEATURE
ANNOTATION In total, we used 108 features to train our model (Supplementary Data 1). In terms of functional features, we annotated domains affected by pLoF variants with PFAM and SMART
domain information. The 3D structure of a protein is essential for proper folding and function of proteins. Therefore, we incorporated two structure-based features, SCOP domains, and
disordered residues, into our pipeline. In addition, we annotated signal peptide and trans-membrane domains. PFAM, SCOP, signal peptide, and trans-membrane domain annotations were obtained
by querying Ensembl Release 73 using the Ensembl PERL API57. Post-translationally modified residues (phosphorylated, acetylated, and ubiquitinated sites) are annotated based on data from
PhosphositePlus25. Disordered residues are known to be important in protein–protein interaction surfaces and have been implicated in disease-causing mechanisms58, 59. We obtained disordered
residues in proteins using DISOPRED24. For all functional features, we addressed the following questions: (1) does the premature stop variant affect a functional feature? and (2) are
functional, structural, or other domains removed due to truncation? Nonsense-mediated decay (NMD) is a cellular surveillance mechanism whereby transcripts containing premature stop codons
are removed to prevent aberrant transcripts and protein products. NMD can be used as a feature to assess whether a transcript containing a pLoF variant will be functional. We therefore
included NMD prediction as a functional feature and identified transcripts containing pLoF variants as candidates for NMD if the distance of the premature stop from the last exon–exon
junction was >50 base pairs. As network features, we calculated proximity parameters for each pLoF-affected gene that correspond to the number of disease genes directly connected to it in
a protein–protein interaction network. Human protein–protein interaction networks were downloaded from BioGrid60 (the version used is BIOGRID-ORGANISM-Homo_sapiens-3.2.95). Dominant and
recessive disease genes were obtained from lists curated from OMIM61,62,63. Shortest path from a gene to the nearest disease gene in the protein–protein interaction network is also included
in the ALoFT output. The evolutionary features considered by ALoFT include the GERP score of the pLoF variant position. In the case of indels, the mean GERP score is provided. In addition,
ALoFT evaluates the evolutionary conservation of the region that is lost due to the truncation. This is calculated as the percentage of coding region lost that occurs in GERP-constrained
elements. d_N_/d_S_ values for human-macaque and human-mouse orthologs were obtained from Ensembl using Biomart. ALoFT also includes all annotation features derived from VAT. This includes
transcript-specific annotation of the coding SNP. In addition, ALoFT provides allele frequency information for the variants based on reference population studies. Specifically, ALoFT output
includes allele frequency information for LoF variants from the Phase1 of 1000 Genomes Project (1KG), ESP6500, as well as ExAC data sets. 1KG includes genetic variation data obtained from
whole genome and exome sequencing of 1092 healthy individuals. ESP6500 consists of genetic variants from exome sequencing of a cohort of 2203 African-American and 4300 European-American
unrelated individuals enrolled in the National Heart, Lung, and Blood Institute Exome Sequencing Project3. The ESP6500 data set was downloaded from Exome Variant Server, NHLBI GO Exome
Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/) (8 November 2013). Version 0.3 of the ExAC data set was downloaded from http://exac.broadinstitute.org/,
containing 60,706 unrelated individuals sequenced as part of various disease-specific and population genetic studies30. An overview of all the features output by ALoFT is shown in
Supplementary Table 1. To account for gene conservation, we calculated synonymous and non-synonymous SNP density based on variation data from 1KG, average GERP scores of synonymous and
non-synonymous SNPs, the percentage of synonymous and non-synonymous SNPs in GERP-constrained elements, the percentage of coding transcript overlapping with constrained GERP elements, and
average heterozygosity for synonymous and non-synonymous SNPs in 1KG. Gene centrality scores were obtained for various networks from Khurana et al.64 Transcript expression levels in 25
tissues from GTex65. For each transcript, we calculated the average expression values across individuals for a particular tissue. Tissue specificity is calculated using a Shannon
entropy-based method66. Number of validated miRNA binding sites per gene were obtained from miRWalk67. Average heterozygosity was calculated as \(\frac{{{\sum} {2pq} }}{l}\), where _p_ is
minor allele frequency, _q_ is the reference allele frequency, _l_ is the length of the coding transcript. ACCOUNTING FOR ANNOTATION ERRORS AND MISMAPPING ERRORS In order to reduce
mismapping errors, ALoFT flags potential false-positive variant calls by identifying homologous regions in the genome where the potential for mismapping is high. This includes variants in
segmentally duplicated regions, variants in genes that have paralogs, and variants in genes that have pseudogenes. 51,599 regions of the human genome are annotated as segmentally duplicated
regions that are at least 1 kb in length and whose sequences are >90% identical. Paralogs of human genes were obtained from Ensembl, with 11,658 genes having paralogs. Pseudogene
information was derived from the GENCODE pseudogene resource68. 3392 genes have pseudogenes. Variants that lead to a premature Stop codon, indels that lead to frameshift and variants in
splice sites are annotated as pLoF variants based on sequence annotation and are assumed to lead to loss of function. However, this assumption is not always valid. Categories of LoF
annotation errors have been evaluated and elucidated in the first systematic catalog of loss-of-function genes2. Thus, the various ways that an inferred LoF annotation may be incorrect are
captured by ALoFT using several flags. _lof_anc_: indicates that the pLoF variant allele is the same as the ancestral allele. Evolutionarily conserved alleles imply that they are likely to
be biologically important and thus represent functional alleles. Therefore, when the pLoF variant is same as the ancestral allele, we believe that it is a functional allele. _near_start_:
The variant is in the first 5% of the coding sequence. _near_end_: The variant is in the last 5% of the coding sequence. _alt_canonical_site_: SNPs in splice sites are flagged as potentially
not LoF when the alternate allele represents the canonical splice site (i.e., when the alternate allele is GT at the donor or AG at the acceptor site). _noncanonical_splice_flank_: variants
in exons that are flanked by noncanonical splice sites. Some of these exons could be due to spurious exon annotations in the gene models. _Small_intron_: variants in introns <15 bp long.
PATHOGENICITY PREDICTION FOR PLOF MUTATIONS To predict the pathogenicity of pLoF variants, we trained a random forest model to differentiate between benign, heterozygous, and homozygous
disease-causing premature stop variants. For the training data, we only used premature stop variants caused by single-nucleotide polymorphisms because indel calling methods are not yet
robust. The benign variant set includes homozygous variants from 1KG. Premature stop mutations leading to disease were obtained from HGMD. To minimize errors due to mistakes in HGMD, we only
used high-confidence mutations labeled as “DM” (disease-causing mutations) in HGMD. We used the variation and gene-specific features that are output by ALoFT to build the classifier. We
also included gene/transcript-specific features, which take into account the effects of length and the background mutation rate for each gene. As training data, we identified benign
premature stop variants as SNPs that are homozygous in at least one individual in 1KG. Premature stop mutations from HGMD are classified as those causing recessive or dominant disease based
on ‘recessive’ and ‘dominant’ genes curated from the Online Mendelian Inheritance in Man database, OMIM61, 62. The training data consist of variants from autosomes only. Mutations that lead
to dominant inheritance of diseases can do so both via loss-of-function mechanisms as well as gain-of-function mechanisms. However, it is reasonable to assume that most pLoF variants in
dominant disease genes cause loss of function. Nonetheless, we only included dominant genes predicted to be haplo-insufficient22 in the training data to make sure that we are predominantly
probing loss-of-function effects. The final training data set was derived from 397 benign premature stop variants (in 380 genes), 3300 dominant premature stop variants (in 136 genes), and
5342 recessive premature stop mutations (in 796 genes) (Supplementary Table 2). In order to classify loss-of-function mutations, descriptive features are transformed into binary values - −1”
and “1”, e.g., whether or not truncating a PFAM domain. Missing values are replaced with the weighted average of the three prediction classes. We then use a random forest algorithm to train
our model and evaluated the performance with 10-fold cross-validations. To reduce bias, we included only one variant per gene in the training data for the benign and recessive classes. The
average number of dominant mutations per gene is 24 (Supplementary Table 2). Therefore, we randomly selected three variants per gene for the dominant class in order to obtain a reasonably
balanced training data set. The variant is picked randomly from the list of mutations and the longest affected transcript is used. Thus, each training model was based on 380 benign premature
stop variants, ~341 dominant mutations, and 796 recessive mutations. Stratified sampling is used in the random forest model to achieve balanced three-class training. We repeated this
process 40 times. We calculated multi-class AUC for the test set using the methodology developed by Hand and Till69. We assigned the class with the highest probability as the predicted
outcome. CLASSIFIER PERFORMANCE EVALUATION In cases where ALoFT returns a similar probability of classification between classes, there is uncertainty in the predicted class. By calculating
the standard deviation of class probabilities across our 40 trained random forest models, we obtain a 95% confidence interval for ALoFT predictions. If the confidence interval of the
predicted class probability overlaps with the confidence interval of either of the two less likely classifications (single-sided test), we attach the label ‘Low Confidence’ (_p_ > 0.05)
to the prediction. Otherwise the prediction is labeled ‘High Confidence’ (_p_ < 0.05). Supplementary Fig. 5 shows the precision calculations for 5 out of the 40 training models. Precision
is calculated as the fraction of true positives among predictions. As an example, for recessive predictions, we counted the number of correct predictions as true positives, the rest of the
recessive predictions as false positives. $${\rm{Precision = }}\frac{{{\rm{True}}\,{\rm{positives}}}}{{{\rm{True}}\,{\rm{positives + False}}\,{\rm{positives}}}}$$ Recall is calculated as:
$${\rm{Recall = }}\frac{{{\rm{True}}\,{\rm{positives}}}}{{{\rm{True}}\,{\rm{positives + False}}\,{\rm{negatives}}}}$$ We evaluated the robustness of the classifier by using several different
training data sets for the prediction. The classifier performs well for all the training data sets as shown in Supplementary Table 3. Olfactory receptor genes have many pseudogenes and
accumulate many LoF mutations70. Therefore, the training data for benign pLoF variants have a higher proportion of high-frequency pLoF variants from this class of genes. In order to avoid
any potential bias arising due to this factor, we validated the robustness of our model by excluding olfactory receptors. Similarly, we show that the model performs well whether we choose
variants from the longest isoform of a gene for the training data or choose any one of the isoforms of the gene. In addition to LoF effects, truncating mutations can also lead to gain of
function. However, gain-of-function mutations are difficult to model systematically as the effect of a variant is very context dependent. In order to minimize errors that might arise due to
inadequate modeling of gain-of-function effects and focus only on LoF, we use predicted haploinsufficient genes as the dominant training set in the final model. However, we show that even a
model where the training data for the dominant class is derived from all dominant genes, the prediction is robust. DETERMINING FEATURE IMPORTANCE In Supplementary Fig. 2, the importance of a
feature is calculated by evaluating the decrease in mean accuracy of the test set when the value of the feature is randomly permuted. The importance plot is not directly interpretable
because some of the prediction variables are correlated. The description of the features can be found in Supplementary Data 1. To further evaluate the features important for the
classification, we built several prediction models using different sets of features for the training. Supplementary Table 4 shows the features used for prediction and their corresponding
multi-class AUC of the test set. APPLICATION OF ALOFT TO SEQUENCING STUDY DATA We applied our method to classify Mendelian pathogenic mutations discovered in the Center For Mendelian
Genomics studies (CMG)33. After excluding training variants, there are 3 dominant and 5 recessive premature stop mutations. We also obtained GERP and CADD32 scores for these variants (Fig.
2b). ClinVar31 variants were obtained from https://github.com/macarthur-lab/clinvar. In order to validate ALoFT predictions, we first excluded all ClinVar variants in genes that were used in
the training set. We then labeled the remaining ClinVar variants as those leading to disease via the dominant or recessive mode of inheritance using an orthogonal list of dominant/recessive
genes obtained from Berg et al.71 To avoid potential bias that might arise due to enrichment of disease variants in particular genes, we randomly picked one variant per gene for the
analysis shown in Fig. 2a. The final set used to validate ALoFT contains 197 variants in genes known to cause disease through the dominant mode of inheritance and 111 variants in recessive
genes. We collected de novo premature stop mutations from four autism studies34,35,36,37. There are 19 and 53 mutations in siblings and probands, respectively. Most individuals have one de
novo premature stop mutation (Supplementary Table 5). The prediction results are included in Supplementary Data 2 (2 out of 53 proband mutations overlap our training data and are excluded in
Fig. 3a). We obtained the list of 33 confident autism genes (FDR < 0.1) from Rubeis et al.39 and observed that dominant disease-causing score for premature stop variants in these genes
are significantly higher than those in other genes (Only de novo pLoFs in probands are used; _p_-value: 5e−3; Wilcoxon rank-sum test; Fig. 3b). We obtained somatic premature stop mutations
from Alexandrov et al.40. This includes 6535 exomes in 30 different cancer types. Cancer genes are from the COSMIC cancer gene consensus72. We used ALoFT as a tool to distinguish passenger
vs. driver mutations in tumors with high mutation burden. For this evaluation, we used ALoFT to identify deleterious LoF mutations. We calculated the ratio of deleterious LoF mutations to
total pLoF mutations for the 6535 exome samples. We binned patient samples with at least one deleterious LoF mutation according to total mutational burden. We applied our method to classify
premature stop variants in the healthy cohort of 1092 individuals from the 1KG data. Among the 5495 premature stop variants (excluding chrX), 148, 3070, and 2277 variants are predicted as
dominant, recessive, and tolerant, respectively (Supplementary Data 4). ESTIMATING LOF MUTATION BURDENING In order to estimate the burden of deleterious LoFs in an individual genome, we
calculated the average number of premature stop variants predicted to be deleterious by ALoFT using data from 1KG Phase 1, 1KG Phase 3, and ExAC. Numerous confounding factors make it
difficult to compare genetic variation data from resequencing studies. For example, the accuracy of variant calls varies depending on the sequencing depth, and different data sets use
different variant calling algorithms and different metrics to evaluate the quality of variants resulting in differing sensitivity and specificity of variant calls. Also, whole-genome
sequencing and whole-exome sequencing provide different genomic coverage, and among exome sequencing studies, different exome capture platforms may have different definitions of exome and
different target enrichment efficiency. 1KG data consist of data obtained both based on exome capture as well as whole-genome sequencing, whereas ExAC is based on exome capture data. To
minimize errors arising from the above-mentioned factors, we used a filtering approach described below to obtain a conservative estimate of the burden of deleterious premature stop variants
in an individual genome. We used high-confidence variants for the calculation of per individual statistics for 1000 Genomes as described below. (1) While ALoFT provides several flags that
identify likely false positive variant calls arising due to mismapping and annotation errors, we conservatively excluded only those pLoF variants that correspond to the ancestral allele as
they are unlikely to result in loss of function. (2) Variants present at >12 alleles (~1% frequency for phase 1 and ~0.5% for phase 3) in either the European or African-American
population of the 1KG cohort, but absent in the ESP6500 cohort were also removed as likely erroneous calls. (3) For the 1KG Phase 1 set, only variants called from exome sequencing (not
available for Phase 3) were included in order to make a fair comparison with the ESP6500 data that is also based on exome capture. We calculated per individual statistics for predicted
dominant, recessive, and benign premature stop mutations and is shown in Supplementary Table 6 and Supplementary Fig. 6. Per individual calculations are based on 246 individuals of African
ancestry and 379 individuals of European ancestry for 1KG Phase 1; 661 individuals of African ancestry and 503 individuals of European ancestry for 1KG Phase 3. For ExAC per individual
calculation, no filtering was applied as we do not want to remove true variant calls that might be present in this data set due to higher sequence coverage. Furthermore, ExAC contains data
aggregated from several disease exome sequencing projects such as inflammatory bowel disease, GoT2D (Type 2 diabetes) consortium, myocardial infarction genetics consortium etc. and some of
the variants might be true disease-causing variants. Thus, our approach provides a lower estimate of the number of potentially deleterious pLoF variants in healthy individuals based on the
value from 1KG Phase1 calculation. DATA AVAILABILITY The ALoFT software can be downloaded from aloft.gersteinlab.org. All ancillary files needed to run the program are included with this
download and described in the Methods section. All analyzed data have been included as Supplementary Data 1–5. Pre-calculated exome-wide ALoFT scores for all base substitutions that
potentially lead to a premature stop codon can be downloaded for both HG19 and GRCh38 human genome reference from aloft.gersteinlab.org. REFERENCES * Balasubramanian, S. et al. Gene
inactivation and its implications for annotation in the era of personal genomics. _Genes Dev._ 25, 1–10 (2011). Article CAS PubMed PubMed Central Google Scholar * MacArthur, D. G. et
al. A systematic survey of loss-of-function variants in human protein-coding genes. _Science (80-.)_ 335, 823–828 (2012). Article ADS CAS Google Scholar * McVean, G. A. et al. An
integrated map of genetic variation from 1,092 human genomes. _Nature_ 491, 56–65 (2012). Article ADS CAS PubMed Google Scholar * 1000 Genomes Project Consortium. et al. A global
reference for human genetic variation. _Nature_ 526, 68–74 (2015). Article Google Scholar * Sulem, P. et al. Identification of a large set of rare complete human knockouts. _Nat. Genet._
47, 448–452 (2015). Article CAS PubMed Google Scholar * Cohen, J. C., Boerwinkle, E., Mosley, T. H. & Hobbs, H. H. Sequence variations in _PCSK9_, low LDL, and protection against
coronary heart disease. _N. Engl. J. Med._ 354, 1264–1272 (2006). Article CAS PubMed Google Scholar * Stein, E. A. et al. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. _N.
Engl. J. Med._ 366, 1108–1118 (2012). Article CAS PubMed Google Scholar * Blom, D. J. et al. A 52-week placebo-controlled trial of evolocumab in hyperlipidemia. _N. Engl. J. Med._ 370,
1809–1819 (2014). Article CAS PubMed Google Scholar * Flannick, J. et al. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. _Nat. Genet._ 46, 357–363 (2014). Article
CAS PubMed PubMed Central Google Scholar * Lim, E. T. et al. Distribution and medical impact of loss-of-function variants in the Finnish founder population. _PLoS Genet._ 10, e1004494
(2014). Article PubMed PubMed Central Google Scholar * Pollin, T. I. et al. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection.
_Science (80-.)_ 322, 1702–1705 (2008). Article ADS CAS Google Scholar * Yu, B. et al. Association of rare loss-of-function alleles in HAL, serum histidine: levels and incident coronary
heart disease. _Circ. Cardiovasc. Genet._ 8, 351–355 (2015). Article CAS PubMed PubMed Central Google Scholar * Stenson, P. D. et al. The human gene mutation database: building a
comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. _Hum. Genet._ 133, 1–9 (2014). Article CAS PubMed Google
Scholar * Isken, O. & Maquat, L. E. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. _Genes Dev._ 21, 1833–1856 (2007). Article CAS PubMed Google
Scholar * Lappalainen, T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. _Nature_ 501, 506–511 (2013). Article ADS CAS PubMed PubMed Central Google
Scholar * Guo, Y. et al. Dissecting disease inheritance modes in a three-dimensional protein network challenges the "guilt-by-association" principle. _Am. J. Hum. Genet._
93, 78–89 (2013). Article CAS PubMed PubMed Central Google Scholar * Adzhubei, I., Jordan, D. M. & Sunyaev, S. R. in _Current Protocols in Human Genetics_ Chapter 7, 7.20.1–7.20.41
(John Wiley & Sons, Inc., 2013). * Cooper, G. M. & Shendure, J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. _Nat. Rev. Genet._ 12,
628–640 (2011). Article CAS PubMed Google Scholar * Karchin, R. Next generation tools for the annotation of human SNPs. _Brief. Bioinform._ 10, 35–52 (2008). Article Google Scholar *
Hu, J. & Ng, P. C. Predicting the effects of frameshifting indels. _Genome Biol._ 13, R9 (2012). Article CAS PubMed PubMed Central Google Scholar * Rausell, A. et al. Analysis of
stop-gain and frameshift variants in human innate immunity genes. _PLoS Comput. Biol._ 10, e1003757 (2014). Article PubMed PubMed Central Google Scholar * Letunic, I., Doerks, T. &
Bork, P. SMART: recent updates, new developments and status in 2015. _Nucleic Acids Res._ 43, D257–D260 (2015). Article CAS PubMed Google Scholar * Finn, R. D. et al. Pfam: the protein
families database. _Nucleic Acids Res._ 42, D222–D230 (2014). Article CAS PubMed Google Scholar * Ward, J. J., McGuffin, L. J., Bryson, K., Buxton, B. F. & Jones, D. T. The DISOPRED
server for the prediction of protein disorder. _Bioinformatics_ 20, 2138–2139 (2004). Article CAS PubMed Google Scholar * Hornbeck, P. V. et al. PhosphoSitePlus: a comprehensive resource
for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. _Nucleic Acids Res._ 40, D261–D270 (2012). Article CAS PubMed
Google Scholar * Cooper, G. M. et al. Distribution and intensity of constraint in mammalian genomic sequence. _Genome Res._ 15, 901–913 (2005). Article CAS PubMed PubMed Central
Google Scholar * Davydov, E. V. et al. Identifying a high fraction of the human genome to be under selective constraint using GERP+ +. _PLoS Comput. Biol._ 6, e1001025 (2010). Article
PubMed PubMed Central Google Scholar * Huang, N., Lee, I., Marcotte, E. M. & Hurles, M. E. Characterising and predicting haploinsufficiency in the human genome. _PLoS Genet._ 6,
e1001154 (2010). Article PubMed PubMed Central Google Scholar * Breiman, L. Random forests. _Mach. Learn._ 45, 5–32 (2001). Article MATH Google Scholar * Lek, M. et al. Analysis of
protein-coding genetic variation in 60,706 humans. _Nature_ 536, 285–291 (2016). Article CAS PubMed PubMed Central Google Scholar * Landrum, M. J. et al. ClinVar: public archive of
relationships among sequence variation and human phenotype. _Nucleic Acids Res._ 42, D980–D985 (2014). Article CAS PubMed Google Scholar * Kircher, M. et al. A general framework for
estimating the relative pathogenicity of human genetic variants. _Nat. Genet._ 46, 310–315 (2014). Article CAS PubMed PubMed Central Google Scholar * Chong, J. X. et al. The genetic
basis of mendelian phenotypes: discoveries, challenges, and opportunities. _Am. J. Hum. Genet._ 97, 199–215 (2015). Article CAS PubMed PubMed Central Google Scholar * Iossifov, I. et
al. De novo gene disruptions in children on the autistic spectrum. _Neuron_ 74, 285–299 (2012). Article CAS PubMed PubMed Central Google Scholar * Neale, B. M. et al. Patterns and rates
of exonic de novo mutations in autism spectrum disorders. _Nature_ 485, 242–245 (2012). Article ADS CAS PubMed PubMed Central Google Scholar * Sanders, S. J. et al. De novo mutations
revealed by whole-exome sequencing are strongly associated with autism. _Nature_ 485, 237–241 (2012). Article ADS CAS PubMed PubMed Central Google Scholar * O’Roak, B. J. et al.
Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. _Nature_ 485, 246–250 (2012). Article ADS PubMed PubMed Central Google Scholar * Jacquemont,
S. et al. A higher mutational burden in females supports a "female protective model" in neurodevelopmental disorders. _Am. J. Hum. Genet._ 94, 415–425 (2014). Article CAS
PubMed PubMed Central Google Scholar * De Rubeis, S. et al. Synaptic, transcriptional and chromatin genes disrupted in autism. _Nature_ 515, 209–215 (2014). Article PubMed PubMed
Central Google Scholar * Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. _Nature_ 500, 415–421 (2013). Article CAS PubMed PubMed Central Google Scholar *
Vogelstein, B. et al. Cancer genome landscapes. _Science (80-.)_ 339, 1546–1558 (2013). Article ADS CAS Google Scholar * Inoue, K. et al. Molecular mechanism for distinct neurological
phenotypes conveyed by allelic truncating mutations. _Nat. Genet._ 36, 361–369 (2004). Article CAS PubMed Google Scholar * Bell, C. J. et al. Carrier testing for severe childhood
recessive diseases by next-generation sequencing. _Sci. Transl. Med._ 3, 65ra4–65ra4 (2011). Article CAS PubMed PubMed Central Google Scholar * Chong, J. X., Ouwenga, R., Anderson, R.
L., Waggoner, D. J. & Ober, C. A population-based study of autosomal-recessive disease-causing mutations in a founder population. _Am. J. Hum. Genet._ 91, 608–620 (2012). Article CAS
PubMed PubMed Central Google Scholar * Cooper, D. N. et al. Genes, mutations, and human inherited disease at the dawn of the age of personalized genomics. _Hum. Mutat._ 31, 631–655
(2010). Article CAS PubMed Google Scholar * Xue, Y. et al. Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and
population-scale resequencing. _Am. J. Hum. Genet._ 91, 1022–1032 (2012). Article CAS PubMed PubMed Central Google Scholar * Tabor, H. K. et al. Pathogenic variants for mendelian and
complex traits in exomes of 6,517 European and African Americans: implications for the return of incidental results. _Am. J. Hum. Genet._ 95, 183–193 (2014). Article CAS PubMed PubMed
Central Google Scholar * Sekiya, A. et al. Compound heterozygotes for filaggrin gene mutations do not always show severe atopic dermatitis. _J. Eur. Acad. Dermatol. Venereol._ 31, 158–162
(2017). Article CAS PubMed Google Scholar * Narasimhan, V. M. et al. Health and population effects of rare gene knockouts in adult humans with related parents. _Science (80-.)_ 352,
474–477 (2016). Article ADS CAS Google Scholar * Austin, E. D. et al. Truncating and missense BMPR2 mutations differentially affect the severity of heritable pulmonary arterial
hypertension. _Respir. Res._ 10, 87 (2009). Article PubMed PubMed Central Google Scholar * Alazami, A. M. et al. Accelerating novel candidate gene discovery in neurogenetic disorders via
whole-exome sequencing of prescreened multiplex consanguineous families. _Cell Rep._ 10, 148–161 (2015). Article CAS PubMed Google Scholar * Alkuraya, F. S. Human knockout research: new
horizons and opportunities. _Trends Genet._ 31, 108–115 (2015). Article CAS PubMed Google Scholar * Yizhak, K., Gabay, O., Cohen, H. & Ruppin, E. Model-based identification of drug
targets that revert disrupted metabolism and its application to ageing. _Nat. Commun._ 4, 2632 (2013). Article ADS PubMed Google Scholar * Bhuvanagiri, M. et al. 5-azacytidine inhibits
nonsense-mediated decay in a MYC-dependent fashion. _EMBO Mol. Med._ 6, 1593–1609 (2014). Article CAS PubMed PubMed Central Google Scholar * Welch, E. M. et al. PTC124 targets genetic
disorders caused by nonsense mutations. _Nature_ 447, 87–91 (2007). Article ADS CAS PubMed Google Scholar * Habegger, L. et al. VAT: a computational framework to functionally annotate
variants in personal genomes within a cloud-computing environment. _Bioinformatics_ 28, 2267–2269 (2012). Article CAS PubMed PubMed Central Google Scholar * Flicek, P. et al. Ensembl
2013. _Nucleic Acids Res._ 41, D48–D55 (2013). Article CAS PubMed Google Scholar * Vacic, V. et al. Disease-associated mutations disrupt functionally important regions of intrinsic
protein disorder. _PLoS Comput. Biol._ 8, e1002709 (2012). Article CAS PubMed PubMed Central Google Scholar * Dunker, A. K. & Obradovic, Z. The protein trinity--linking function and
disorder. _Nat. Biotechnol._ 19, 805–806 (2001). Article CAS PubMed Google Scholar * Stark, C. et al. BioGRID: a general repository for interaction datasets. _Nucleic Acids Res._ 34,
D535–D539 (2006). Article CAS PubMed Google Scholar * Hamosh, A., Scott, A. F., Amberger, J. S., Bocchini, C. A. & McKusick, V. A. Online mendelian inheritance in man (OMIM), a
knowledgebase of human genes and genetic disorders. _Nucleic Acids Res._ 33, D514–D517 (2004). Article PubMed Central Google Scholar * Blekhman, R. et al. Natural selection on genes that
underlie human disease susceptibility. _Curr. Biol._ 18, 883–889 (2008). Article CAS PubMed PubMed Central Google Scholar * Boone, P. M. et al. Deletions of recessive disease genes: CNV
contribution to carrier states and disease-causing alleles. _Genome Res._ 23, 1383–1394 (2013). Article CAS PubMed PubMed Central Google Scholar * Khurana, E. et al. Integrative
annotation of variants from 1092 humans: application to cancer genomics. _Science_ 342, 1235587 (2013). Article PubMed PubMed Central Google Scholar * Lonsdale, J. et al. The
genotype-tissue expression (GTEx) project. _Nat. Genet._ 45, 580–585 (2013). Article CAS Google Scholar * Schug, J. et al. Promoter features related to tissue specificity as measured by
Shannon entropy. _Genome Biol._ 6, R33 (2005). Article PubMed PubMed Central Google Scholar * Dweep, H., Sticht, C., Pandey, P. & Gretz, N. miRWalk—database: prediction of possible
miRNA binding sites by ‘walking’ the genes of three genomes. _J. Biomed. Inform._ 44, 839–847 (2011). Article CAS PubMed Google Scholar * Pei, B. et al. The GENCODE pseudogene resource.
_Genome Biol._ 13, R51 (2012). Article CAS PubMed PubMed Central Google Scholar * Hand, D. J. & Till, R. J. A simple generalisation of the area under the ROC curve for multiple
class classification problems. _Mach. Learn._ 45, 171–186 (2001). Article MATH Google Scholar * Glusman, G., Yanai, I., Rubin, I. & Lancet, D. The complete human olfactory subgenome.
_Genome Res._ 11, 685–702 (2001). Article CAS PubMed Google Scholar * Berg, J. S. et al. An informatics approach to analyzing the incidentalome. _Genet. Med._ 15, 36–44 (2013). Article
CAS PubMed Google Scholar * Forbes, S. A. et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. _Nucleic Acids Res._ 43, D805–D811 (2015). Article CAS
PubMed Google Scholar Download references ACKNOWLEDGEMENTS We thank Daniel Spakowicz for comments on the manuscript. This work was supported by grants 5R01GM104371 (US National Institutes
of Health/National Institute of General Medical Sciences) to S.B. and D.G.M., and U54HG006504 (Yale Center for Mendelian Genomics) to M.G. AUTHOR INFORMATION Author notes * Suganthi
Balasubramanian Present address: Regeneron Genetics Center, Tarrytown, NY, 10591, USA * Yao Fu Present address: Bina Technologies, Part of Roche Sequencing, Belmont, CA, 94002, USA *
Suganthi Balasubramanian and Yao Fu contributed equally to this work. AUTHORS AND AFFILIATIONS * Program in Computational Biology and Bioinformatics, Yale University, New Haven, CT, 06520,
USA Suganthi Balasubramanian, Yao Fu & Mark Gerstein * Molecular Biophysics and Biochemistry Department, Yale University, New Haven, CT, 06520, USA Suganthi Balasubramanian, Mayur
Pawashe, Patrick McGillivray, Mike Jin, Jeremy Liu & Mark Gerstein * Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, 02114, USA Konrad J. Karczewski
& Daniel G. MacArthur * Program in Medical and Population Genetics, Broad Institute of MIT and Harvard, Cambridge, Massachusetts, 02142, USA Konrad J. Karczewski & Daniel G.
MacArthur * Department of Computer Science, Yale University, New Haven, CT, 06520, USA Mark Gerstein Authors * Suganthi Balasubramanian View author publications You can also search for this
author inPubMed Google Scholar * Yao Fu View author publications You can also search for this author inPubMed Google Scholar * Mayur Pawashe View author publications You can also search for
this author inPubMed Google Scholar * Patrick McGillivray View author publications You can also search for this author inPubMed Google Scholar * Mike Jin View author publications You can
also search for this author inPubMed Google Scholar * Jeremy Liu View author publications You can also search for this author inPubMed Google Scholar * Konrad J. Karczewski View author
publications You can also search for this author inPubMed Google Scholar * Daniel G. MacArthur View author publications You can also search for this author inPubMed Google Scholar * Mark
Gerstein View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS S.B., Y.F. and M.G. contributed to the design of the study. M.P. wrote the
software and built the aloft.gersteinlab.org website. S.B., Y.F. and P.M. performed data analyses and interpretation. M.J. and J.L. contributed to early versions of the code base. K.J.K. and
D.G.M. provided per sample analysis data for the ExAC cohort. D.G.M. edited an early version of the manuscript and provided useful feedback. S.B., Y.F., P.M. and M.G. wrote the manuscript.
CORRESPONDING AUTHORS Correspondence to Suganthi Balasubramanian or Mark Gerstein. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. ADDITIONAL
INFORMATION PUBLISHER'S NOTE: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. ELECTRONIC SUPPLEMENTARY MATERIAL
SUPPLEMENTARY INFORMATION SUPPLEMENTARY DATA 1 SUPPLEMENTARY DATA 2 SUPPLEMENTARY DATA 3 SUPPLEMENTARY DATA 4 SUPPLEMENTARY DATA 5 RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed
under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give
appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in
this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative
Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a
copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Balasubramanian, S., Fu, Y., Pawashe, M. _et al._ Using
ALoFT to determine the impact of putative loss-of-function variants in protein-coding genes. _Nat Commun_ 8, 382 (2017). https://doi.org/10.1038/s41467-017-00443-5 Download citation *
Received: 20 May 2016 * Accepted: 29 June 2017 * Published: 29 August 2017 * DOI: https://doi.org/10.1038/s41467-017-00443-5 SHARE THIS ARTICLE Anyone you share the following link with will
be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt
content-sharing initiative