Play all audios:
ABSTRACT Rosacea is a chronic inflammatory skin disorder, whose underlying cellular and molecular mechanisms remain obscure. Here, we generate a single-cell atlas of facial skin from female
rosacea patients and healthy individuals. Among keratinocytes, a subpopulation characterized by IFNγ-mediated barrier function damage is found to be unique to rosacea lesions. Blocking IFNγ
signaling alleviates rosacea-like phenotypes and skin barrier damage in mice. The papulopustular rosacea is featured by expansion of pro-inflammatory fibroblasts, Schwann, endothelial and
macrophage/dendritic cells. The frequencies of type 1/17 and skin-resident memory T cells are increased, and vascular mural cells are characterized by activation of inflammatory pathways and
impaired muscle contraction function in rosacea. Most importantly, fibroblasts are identified as the leading cell type producing pro-inflammatory and vasodilative signals in rosacea.
Depletion of fibroblasts or knockdown of PTGDS, a gene specifically upregulated in fibroblasts, blocks rosacea development in mice. Our study provides a comprehensive understanding of the
aberrant alterations of skin-resident cell populations and identifies fibroblasts as a key determinant in rosacea development. SIMILAR CONTENT BEING VIEWED BY OTHERS ACSL5 MEDIATES
MACROPHAGE INFILTRATION AND LIPID METABOLISM IN ERYTHROTELANGIECTASIA ROSACEA VIA POTENTIAL PATHOGENIC MECHANISMS AND THERAPEUTIC TARGETS Article Open access 08 April 2025 OTULIN MAINTAINS
SKIN HOMEOSTASIS BY CONTROLLING KERATINOCYTE DEATH AND STEM CELL IDENTITY Article Open access 08 October 2021 SINGLE CELL TRANSCRIPTIONAL ZONATION OF HUMAN PSORIASIS SKIN IDENTIFIES AN
ALTERNATIVE IMMUNOREGULATORY AXIS CONDUCTED BY SKIN RESIDENT CELLS Article Open access 06 May 2021 INTRODUCTION Rosacea is a commonly chronic inflammatory skin disorder that mainly affects
the central face. The prevalence has been estimated to range from less than 1 to 22% across populations worldwide1,2,3. Rosacea is characterized by facial erythema, telangiectasia, edema,
papules, pustules and recurrent flushing. According to the clinical features, it is classified into four subtypes, including erythematotelangiectatic (ETR), papulopustular (PPR), phymatous
(PhR), and ocular rosacea, although there may be overlaps between different subtypes4,5. The pathogenesis involves damaged skin barrier, dysregulated inflammatory responses (perivascular or
pilosebaceous infiltration), hyperactive neurovascular reactivity (dilation), glandular hyperplasia and fibrotic changes6,7,8,9,10,11, a composition reflecting the multivariate process of
this disease. To date, existing concepts regarding the pathophysiology of rosacea have been obscure and mainly limited to histomorphology, global gene analysis and immunohistochemistry due
to the fact that laboratory models are not fully representative of rosacea7,12,13,14,15,16. Single-cell RNA-sequencing (scRNA-seq) has emerged as a powerful tool to define the cellular
composition (including novel or rare cell subpopulations), cell-specific transcriptome dynamics, cell lineage tracing and cell-cell communications in complex tissues at uniquely high
resolution17,18,19,20. However, to our knowledge, a study regarding scRNA-seq application for investigating the pathogenesis of rosacea has never been reported. Here, we report scRNA-seq
analysis for 131,243 cells from 3 conditions: lesional and non-lesional facial skin from female rosacea patients, and facial skin from female healthy individuals. Via single-cell-level
resolution, we provide an atlas of the complexity and diversity of aberrant skin-resident cell populations in rosacea. Most importantly, by combining cell-cell communication analysis with
mouse model assays, we identify fibroblasts as a key determinant in the pathogenesis of rosacea, suggesting an important role for mesenchymal subpopulations in the treatment of inflammatory
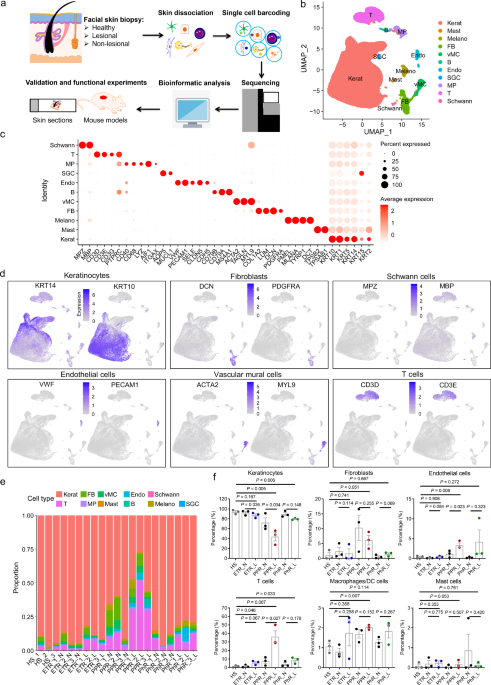
skin disorders. RESULTS SCRNA-SEQ ATLAS OF CELL POPULATIONS IN ROSACEA We obtained skin biopsies from lesional and non-lesional facial skin of 9 female rosacea patients (including 3 ETR, 3
PPR and 3 PhR) and facial skin of 3 female healthy individuals. To dissect the cellular and molecular changes in rosacea, we performed scRNA-seq on these skin biopsies from the seven
conditions: facial skin from healthy individuals (HS), non-lesional and lesional facial skin from ETR patients (ETR_N and ETR_L), non-lesional and lesional facial skin from PPR patients
(PPR_N and PPR_L), non-lesional and lesional facial skin from PhR patients (PhR_N and PhR_L) (Fig. 1a). After quality control (“Methods”), 131,243 cells were reserved for subsequent
analyses. Using the integration methods from Seurat R package, total 28 clusters (c0-c27) were obtained, which were distinguished by distinct marker genes (Supplementary Fig. 1a). These
clusters could be reproduced with cells from each of the 21 samples, suggesting that they are robustly present across different samples and conditions (including HS, ETR_N, ETR_L, PPR_N,
PPR_L, PhR_N and PhR_L) (Supplementary Fig. 1b, c). The above 28 clusters were further identified as 11 different cell types, based on the expression of known markers, including
keratinocytes (KRT14+, KRT10+), fibroblasts (DCN+, PDGFRA+), Schwann cells (MPZ+, MBP+), endothelial cells (VWF+, PECAM1+), vascular mural cells (vMCs) (ACTA2+, MYL9+), T cells (CD3D+,
CD3E+), macrophages/dendritic cells (DCs) (AIF1+), melanocytes (PMEL+), sweat gland cells (AQP5+), B cells (CD79A+), mast cells (TPSB2+)21,22,23,24,25 (Supplementary Fig. 2; Fig. 1b–d;
Supplementary Data 1; Supplementary Fig. 3a–e). We defined the relative proportion of each cell type in all samples, showing significant alterations in cellular composition in rosacea skin
compared with HS (Fig. 1e; Supplementary Data 2 and 3). The immune cell infiltrations were made up of T cells, macrophages/dendritic cells, mast cells and B cells in the lesional skin of
rosacea patients. Thereinto, T cells were the dominant infiltrating cells, and increased in the lesional skin of all rosacea subtypes, even in the non-lesional skin of PPR and PhR (HS:
1.398%, ETR_N: 1.586%, ETR_L: 5.767%, PPR_N: 7.536%, PPR_L: 36.241%, PhR_N: 3.97%, PhR_L: 8.682%); macrophages/dendritic cells were significantly increased in the lesional skin of PPR, and
slightly increased in the lesional skin of ETR and PhR (HS: 1.058%, ETR_N: 0.8%, ETR_L: 1.742%, PPR_N: 1.695%, PPR_L: 2.03%, PhR_N: 1.27%, PhR_L: 1.833%); the proportions of mast cells
showed no significant changes; while B cells were mainly increased in the lesional skin of PPR (HS: 0.023%, ETR_N: 0.029%, ETR_L: 0.119%, PPR_N: 0.0365%, PPR_L: 1.749%, PhR_N: 0.019%, PhR_L:
0.219%) (Fig. 1f; Supplementary Fig. 3f). Only PPR skin displayed a slight increase in the proportion of fibroblasts. Endothelial cells were obviously increased in PPR and slightly
increased in PhR (HS: 0.387%, ETR_N: 0.143%, ETR_L: 0.415%, PPR_N: 1.14%, PPR_L: 3.327%, PhR_N: 0.234%, PhR_L: 4.202%) (Fig. 1f). Due to the increase in inflammatory cell infiltrations,
keratinocytes were decreased in the lesional skin of all rosacea types, even in the non-lesional skin of PPR (HS: 93.82%, ETR_N: 92.50%, ETR_L: 87.36%, PPR_N: 72.31%, PPR_L: 43.90%, PhR_N:
89.58%, PhR_L: 79.56%) (Fig. 1f). However, the proportions of other cells, including vascular mural cells, Schwann cells, melanocytes and sweat gland cells, showed no remarkable alterations
(Supplementary Fig. 3f). Since the healthy skin samples were dominated by epidermal cells, this caveat should be kept in mind regarding all conclusions for cellular proportion (especially
for fibroblasts). Taken together, these results reveal the composition and alterations of cell populations in rosacea skin. IDENTIFICATION OF KERATINOCYTE SUBPOPULATIONS IN ROSACEA
Keratinocytes, the most dominant cell type in the epidermis of skin, have been demonstrated to participate in the pathogenesis of multiple inflammatory skin disorders, such as psoriasis and
atopic dermatitis26. Here, by performing subclustering analysis, keratinocytes were separated into 14 subclusters (Supplementary Fig. 4a). According to the structure of human skin and the
expression of known markers27,28, the above clusters were further identified as 4 classical subpopulations, including basal stem (KRT15+), mitotic (MKI67+), spinous (KRT1+, KRT10+), and
granular (FLG+, LOR+) keratinocytes (Fig. 2a; Supplementary Fig. 4b, c; Supplementary Data 4). In addition, we unexpectedly found a keratinocyte subpopulation with high expression of CD74,
which was mainly expanded in the lesional skin of PPR, and slightly increased in ETR and PhR (Fig. 2a–d; Supplementary Fig. 4b; Supplementary Data 4). By immunostaining, we confirmed the
expansion of CD74+ keratinocytes in the lesional epidermis of all rosacea types (Fig. 2e, f). To determine the differences in CD74+ keratinocytes between lesional and healthy or non-lesional
skin, we performed Gene Set Enrichment Analysis (GSEA) using both Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) terms. Our results showed that in CD74+ keratinocytes
of lesional skin, interferon (IFN) related pathways were significantly upregulated, whereas skin barrier-related pathways, such as tight junction, adherens junction and cell adhesion
molecules, were significantly downregulated (Fig. 2g, h). Notably, the identified skin barrier damage in rosacea was characterized by the downregulation of multiple claudins (CLDNs) (Fig.
2i), consistent with our previous report8. The hyperactivation of IFN signaling pathway was further supported by the upregulation of multiple IFN-related genes and by immunostaining of IRF1
and p-STAT1, two active downstream proteins of IFN signaling pathway29 (Fig. 2j–l; Supplementary Fig. 4d, e). Previous studies have linked IFN signaling to epidermal barrier function and the
pathogenesis of various inflammatory skin diseases30,31,32,33. However, the functional role of IFN signaling in rosacea remains largely unknown. We first analyzed the expression of type I
interferons, including IFNα (IFNA1, IFNA2, IFNA4, IFNA5, IFNA6, IFNA7, IFNA8, IFNA10, IFNA13, IFNA14, IFNA16, IFNA17 and IFNA21), IFNβ (IFNB1), IFNω (IFNW1), IFNɛ (IFNE), IFNк (IFNK), and
type II interferon (IFNG) in our scRNA-seq datasets. Our results showed that none of the type I interferons was detected, but type II interferon (IFNG) was obviously and specifically
expressed in T cells and upregulated in the lesional skin of different rosacea subtypes (Supplementary Fig. 5a–g). These results were further confirmed by our previously published bulk
RNA-seq datasets34 (Supplementary Data 5; Supplementary Fig. 5h). Furthermore, we analyzed the expression of type I IFN receptors (IFNAR1 and IFNAR2) and type II IFN receptors (IFNGR1 and
IFNGR2), and showed that both type IFN receptors were expressed in the facial skin, and type II IFN receptors were robustly expressed in epidermal keratinocytes (Supplementary Fig. 5i, j).
All the above results suggest an important role of IFN gamma (IFNγ) signaling in rosacea. To this end, we first generated a cathelicidin LL37-induced rosacea-like mouse model, which
resembles the clinical symptoms of rosacea patients in accordance with previous studies10,16. By co-immunostaining of CD4 and IFNγ, we further confirmed that IFNγ was mainly expressed and
upregulated in the T cells of lesional skin of rosacea-like mouse model (Supplementary Fig. 6a, b). Then, IFNγ neutralizing antibodies were subcutaneously injected to investigate the
functional role of IFNγ signaling in the development of rosacea (Supplementary Fig. 7a). As expected, neutralizing antibody injection significantly inhibited the activation of IFNγ signaling
and obviously alleviated the rosacea-like phenotypes in mice (Supplementary Fig. 7b, c; Fig. 2m, n). The dermal infiltrating cells and rosacea-associated gene expression were also improved
(Supplementary Fig. 7d; Fig. 2o, p). Moreover, we found that neutralization of IFNγ not only inhibited the expansion of CD74+ keratinocytes (Supplementary Fig. 7e, f) but also restored the
expression of multiple epidermal barrier genes (mainly CLDNs) and transepidermal water loss (TEWL) in LL37-induced mice skin (Fig. 2q–t; Supplementary Fig. 7g). Consistently, the expression
of epidermal barrier genes, including CLDNs and tight junction proteins (TJPs), was impaired in IFNγ-treated human keratinocytes in vitro (Supplementary Fig. 7h). Collectively, these results
identify a disease-enriched keratinocyte subpopulation with hyperactivation of IFNγ signaling, which may be responsible for the skin barrier damage and pathogenesis of rosacea.
IDENTIFICATION OF FIBROBLAST SUBPOPULATIONS IN ROSACEA Fibroblasts are the main cell type in the dermis, which have been reported to play different roles in the pathogenesis of inflammatory
diseases35. However, the functional role of fibroblasts in rosacea development is unknown. In this study, by performing subclustering analysis, fibroblasts were separated into 10 subclusters
(Supplementary Fig. 8a). Hierarchical cluster analysis demonstrated that fibroblasts could further be divided into 5 subpopulations, referred to as complement C3+ (C3+), IGFBP3+, SFRP2+,
ASPN+, and DSG1+ fibroblasts according to their specific markers respectively. Among these, C3+ fibroblasts were found to be unique to the lesional skin of PPR rosacea (Supplementary Fig.
8b–e; Fig. 3a–d; Supplementary Data 6). Considering HS_2 and HS_3 samples were dominated by epidermal cells, and only 4 and 9 fibroblast cells were collected respectively (Supplementary Data
3), this caveat should be kept in mind regarding the proportion of fibroblasts. By co-immunostaining of C3 and DCN, a marker of fibroblasts21 (Fig. 1d), we verified the significant
expansion of C3+ fibroblasts in the lesional skin of PPR patients and revealed a slight increase in ETR and PhR lesional skin (Fig. 3e, f). To figure out the potential role of C3+
fibroblasts in the pathogenesis of rosacea, we conducted Gene Set Enrichment Analysis (GSEA) by using the marker genes (Supplementary Data 6). Our results showed that the upregulated
pathways were mainly associated with inflammation, such as chemokine signaling pathway, cytokine-cytokine receptor interaction, TNF signaling pathway and NF-kappa B signaling pathway,
suggesting an apparent pro-inflammatory role for C3+ fibroblasts (Fig. 3g). Consistently, a series of chemokines, including _CCL19, CXCL1, CXCL2_ and _CXCL12_, was demonstrated to be highly
expressed in C3+ fibroblasts in the lesional skin of PPR patients (Fig. 3h, i; Supplementary Data 6). More importantly, we demonstrated that these chemokines (_CCL19, CXCL1, CXCL2_ and
_CXCL12_) were basically expressed in C3+ fibroblasts across all cell types of the whole skin (Supplementary Fig. 8f; Fig. 3j), suggesting they are fibroblast-specific chemokines in rosacea
skin lesions. IFNγ neutralization decreased the expression of CCL19 in the fibroblasts of LL37-induced mice skin (Supplementary Fig. 8g, h). Collectively, these results reveal that
fibroblasts contribute to the amplification of inflammation through transition to a pro-inflammatory state in the pathogenesis of rosacea. IDENTIFICATION OF SCHWANN CELL SUBPOPULATIONS IN
ROSACEA Numerous lines of evidence suggest a key role of neurogenic inflammation in rosacea development11,36,37,38,39. Although any type of neuron cell was not found in our scRNA-seq
datasets, we identified a cluster of Schwann cells, the major glial cell type in the peripheral nervous system. By performing subclustering analysis, Schwann cells were separated into 2
subclusters (Supplementary Fig. 9a, b; Supplementary Data 7). Among these, a CD74+ subpopulation was found to be remarkably expanded in the lesional skin of PPR patients (Supplementary Fig.
9c–e), which was further confirmed by co-immunostaining of CD74 and MBP (Supplementary Fig. 9f, g); and the sample sizes are too small to know if other rosacea subtypes have similar
findings. However, these CD74+ Schwann cells were not found in LL37-induced rosacea-like mice skin (Supplementary Fig. 9h). To illustrate the potential role of Schwann cells in the
development of rosacea, we performed GSEA. Our results demonstrated that the upregulated pathways were primarily involved in inflammation, such as IL-17 signaling pathway, inflammatory
mediator regulation of TRP channels, Toll-like receptor signaling pathway and antigen processing and presentation (Supplementary Fig. 9i), suggesting Schwann cells receive more inflammatory
signals, which may play a role in the pathogenesis of rosacea. CHANGES IN DIFFERENT IMMUNE CELL POPULATIONS IN ROSACEA Dysregulation of the immune system is the hallmark of rosacea9, but the
changes in immune cells at the single-cell level have not been defined. As described above, our results showed that T cells are the dominant infiltrating cells, and macrophages/DCs and B
cells are also increased in the lesional skin of rosacea, especially in PPR (Fig. 1e, f; Supplementary Fig. 3f; Supplementary Data 2 and 3). To investigate the changes of T cells in rosacea,
we first analyzed the transcriptional profile, and found that multiple inflammation-associated genes, including S100As, IFNG and CCL540,41,42, were upregulated in T cells in the lesional
skin of all subtypes of rosacea (Supplementary Fig. 10a). Next, we integrated T cells from all samples and yielded 7 subclusters (T_c0 to T_c6) according to the previously described
strategies25, among which T_c1 is CD8 positive while others are CD4 positive cells (Fig. 4a; Supplementary Fig. 10b). Detailedly, T_c0 (highly expressed Th1 marker genes, _IFNG_ and
_TBX21_43) and T_c2 (highly expressed Th17 marker genes, _IL17A_ and _RORC_44) were obviously increased in the lesional skin of PPR patients, and slightly expanded in certain ETR or PhR
patients; T_c6 (expressed Th2 marker gene, _IL13_45) was also slightly increased. Compared with HS and N skin, T_c4 (highly expressed Treg marker genes, _FOXP3_ and _IL2RA_46) showed an
increase in the lesional skin of PPR and PhR patients (Fig. 4b–d). Notably, we found that T_c3 (highly expressed TRM marker gene, _ITGAE_21) was mildly expanded in rosacea (Fig. 4b–d), which
was confirmed by the co-immunostaining of CD69 and CD103, two typical markers of TRM cells21 (Fig. 4e, f). Macrophages/DCs were separated into 1 macrophage and 3 DC (DC1, DC2 and DC3)
populations (Fig. 4g). Among these, macrophages were identified by expression of _CD68, ITGAM_ and _FCGR1A_ (Supplementary Fig. 10c). Although all DCs expressed AIF1 (Supplementary Fig. 3a),
DC1 and DC3 were characterized by higher expression of _CD1A_ and _FCER1A_ (Supplementary Fig. 10d). Compared with HS and N skin, the percentage of macrophages was significantly expanded in
the lesional skin of PPR and slightly increased in ETR and PhR, while DCs showed no obvious alteration in rosacea (Fig. 4h, i; Supplementary Fig. 10e, f). To uncover the transcriptional
changes of macrophages between lesional skin and HS/N, we performed differentially expressed genes (DEGs) analysis and found that multiple chemokines (including CCL5, CXCL9, CXCL10, CXCL11,
CXCL12) involved in T cell activation and differentiation were increased in the lesional skin of PPR patients (Fig. 4j). CXCL10+ macrophages were also increased in rosacea mouse model, which
was restored by IFNγ neutralization (Supplementary Fig. 10g, h). Further GSEA analysis showed that complement and coagulation cascades, antigen processing and presentation, Toll-like
receptor signaling pathway related to immune response were commonly upregulated in macrophages of all subtypes of rosacea (Fig. 4k). Collectively, these results suggest that Th1/Th17
polarization and macrophage infiltration are the hallmarks of skin inflammation, and resident memory T cells may play an underestimated role in rosacea. ALTERATIONS IN VASCULAR CELL
POPULATIONS IN ROSACEA Besides inflammation, vascular dysfunction, such as vasodilation, is another hallmark of rosacea1. However, the cellular and molecular changes in cells of blood vessel
structures remain obscure. We first analyzed vascular mural cells (vMCs), including vascular smooth muscle cells (vSMCs) and pericytes, which are responsible for the contraction and
relaxation of vascular vessels47,48,49,50. First, according to previous study21, pericytes were distinguished from vSMCs by RGS5, a marker for pericytes (Supplementary Fig. 11a, b). By
performing subclustering analysis, vSMCs were separated into 2 subclusters, but the percentage and distribution of subclusters were comparable across HS, N and L, suggesting that vSMC cell
identity may not alter in rosacea (Supplementary Fig. 11c, d; Supplementary Data 8). To discover the potential changes of vSMCs in rosacea, we performed GSEA analysis and found that multiple
pathways involved in inflammatory responses were routinely upregulated in vSMCs, such as IL-17 signaling pathway (Supplementary Fig. 11e, f). Pericytes were separated into 2 subclusters,
whose percentage and distribution were also not affected in rosacea (Supplementary Fig. 11g, h; Supplementary Data 9). Similar to vSMCs, several inflammatory pathways were activated in
pericytes, including IL-17 signaling pathway and NF-Kappa B signaling pathway (Supplementary Fig. 11i). Although alterations in muscle contraction function were not identified in our
scRNA-seq datasets, we detected the contractile activity of vMCs by co-immunostaining of α-SMA (a vMC marker) and phosphorylated myosin light chain 2 (p-MLC2), an indicator for myosin ATPase
activity and smooth muscle contraction51. Our results showed that the percentage of p-MLC2+ vMCs was dramatically decreased in the lesional skin of all subtypes of rosacea (Supplementary
Fig. 11j, k), but the protein levels of total MLC2 were not affected (Supplementary Fig. 11l–n), revealing a deficiency of muscle contraction in vMCs in rosacea. Next, we analyzed the
changes in endothelial cells, another dominant cell type in blood vessel structures. By UMAP analysis, endothelial cells were separated into 5 subclusters (Supplementary Fig. 12a), among
which subcluster 1 was significantly increased in the lesional skin of rosacea patients (Supplementary Fig. 12b, c). Further GSEA analysis showed that type I interferon signaling pathway was
enriched in this subcluster, suggesting a pro-inflammatory role of endothelial cells in the pathogenesis of rosacea (Supplementary Fig. 12d). FIBROBLASTS ARE IDENTIFIED AS A MAJOR
DETERMINANT IN ROSACEA Since multiple resident cell types exhibited alterations responsible for the clinical manifestations of rosacea patients (such as increased T cell activation and
differentiation for inflammation, and impaired muscle contraction of vMCs for vasodilation), we wondered which cell type plays a dominant role in orchestrating the complicated network to
initiate rosacea. To this end, we performed CellChat analysis52, to investigate the intercellular interactions among cell types in rosacea. The overall interaction number and strength were
both increased in the lesional skin of all rosacea subtypes, especially in PPR (Fig. 5a and Supplementary Fig. 13a). Incoming/outgoing interaction strength results revealed that fibroblasts
were the predominant cells generating outgoing signals, while immune cells like T cells, B cells and macrophages are the predominant recipient cells (Fig. 5b–d). Moreover, macrophages,
endothelial cells, vMCs, keratinocytes and T cells are the primary targets for the outgoing signals of fibroblasts (Supplementary Fig. 13b–d). These results suggest a potentially critical
role of fibroblasts in rosacea development. To further determine the mechanisms by which fibroblasts are involved in the pathogenesis of rosacea, GSEA analysis was performed for each cell
type using the enriched genes (compared to all other cells, Supplementary Data 10) in the lesional skin of rosacea patients. Our results showed that fibroblasts were overrepresented in
categories related to the pro-inflammatory pathways (chemokine signaling pathway and cytokine-cytokine receptor interaction) and vasodilative pathway (relaxin signaling pathway) in all cell
types (Fig. 5e), which was further confirmed by the expression of their signature genes, such as CCL19 and PTGDS (Fig. 5f and Supplementary Data 10). Thereinto, fibroblasts were shown to
express high levels of chemokine CCL19, which was connected by ligand-receptor interactions to CCR7+ T cells mainly in PPR (Fig. 5g–i; Supplementary Fig. 13e). Moreover, as a vasodilation
related factor synthase53,54, PTGDS was specifically expressed in fibroblasts and significantly increased in rosacea, especially in ETR and PPR (Fig. 5j–m; Supplementary Fig. 13f, g). As a
consequence, PGD2, the product of PTGDS and a typical mediator of vasodilation53,54, was significantly increased in the lesional skin of rosacea (Fig. 5n). The functional role of PGD2 was
further supported by the evidence showing that PTGDR, the receptor of PGD2, was highly expressed in vMCs of the skin (Fig. 5o; Supplementary Fig. 13h). Collectively, these results suggest
that the fibroblasts play the key role in cell interactions with immune and vascular mural cells in rosacea development. Given that fibroblasts are identified as the major cell type yielding
pro-inflammatory and vasodilative signals in the lesional skin, we wondered whether these cells play a functional role in the development of rosacea. To this end, we generated mice
transgenic for the diphtheria toxin receptor under the control of the Col1a2 promoter (Col1a2DTR) by mating Col1a2-CreER mice with iDTR mice. Col1a2DTR mice allow for robust depletion of
skin fibroblasts following administration of tamoxifen and diphtheria toxin (DT) (Supplementary Fig. 14a–c). We then intradermally injected cathelicidin LL37 into Col1a2DTR and wildtype (WT)
mice (Supplementary Fig. 14a), to establish rosacea-like mouse models as previously described10,11,16,36. Our results showed that 12 h post the last LL37 injection, WT mice displayed
obvious rosacea-like dermatitis, but skin fibroblast ablation mice were unable to develop typical rosacea-like phenotypes (Fig. 6a–c). Furthermore, the inflammatory cell infiltration in the
dermis and disease-characteristic inflammatory factors were also significantly improved in these mice (Fig. 6d–f). To further verify the critical role of fibroblasts in the pathogenesis of
rosacea, we focused on PTGDS, a gene specifically expressed and upregulated in fibroblasts of rosacea skin lesions (Fig. 5f, j–m). First, PTGDS was demonstrated to be also specifically
expressed and upregulated in fibroblasts of LL37-induced rosacea-like mouse model (Supplementary Fig. 15a, b). To explore the functional significance of PTGDS in rosacea development, we
injected _Ptgds_ and scrambled siRNAs intradermally twice at the indicated time to knockdown Ptgds in mouse skin (Supplementary Fig. 15c). Then, we verified that PTGDS and PGD2 were both
suppressed in LL37-injected skin after _Ptgds_ siRNAs treatment (Supplementary Fig. 15a, b and d). Our results showed that the knockdown of _Ptgds_ not only dramatically improved the
rosacea-like features (Fig. 6g–i), but also decreased the dermal infiltrating cells and inflammatory factors in mice (Fig. 6j, k; Supplementary Fig. 15e). Consistently, CCL19+ fibroblasts
and CCR7+ T cells were abrogated after _Ptgds_ siRNAs administration in LL37-injected skin (Supplementary Fig. 15f–i). Moreover, by immunohistochemistry (IHC) of CD31, we demonstrated that
_Ptgds_ knockdown improved the abnormal dilation of blood vessels (Fig. 6l, m), which was further supported by immunostaining of p-MLC2 (Supplementary Fig. 15j, k). Taken together, these
findings demonstrate that fibroblasts act as a major determinant in the pathogenesis of rosacea. DISCUSSION Although rosacea is considered to be involved in the dysregulation of cutaneous
nervous, immune and vascular systems, the precise cell composition and their molecular alterations and functions in cutaneous lesions in this disorder remain unclear. Here we have presented
what, to the best of our knowledge, is currently the only comprehensive single-cell transcriptomic analysis of all cell types within skin lesions from patients with rosacea compared with
healthy individuals. Our scRNA-seq atlas of rosacea and healthy skin contains 11 broad cell types. By high-resolution subclustering, functional analysis and experimental validation, we
identified multiple subpopulations associated with rosacea in different cell types. Skin barrier is impaired in rosacea skin lesions characterized by decreased expression of multiple barrier
genes, especially CLDNs, which might be responsible for the exacerbation of inflammation7,8,55. Our scRNA-seq data here identify a subpopulation of keratinocytes increased in the lesional
skin of all rosacea subtypes, which is featured by barrier function damage, including declined tight junction, adherens junction and cell adhesion molecules. Furthermore, based on scRNA-seq
data analysis and functional experimentation, we demonstrate that the upregulated type II IFN (IFNγ) signaling might contribute to the impaired skin barrier, especially for the downregulated
CLDNs in keratinocytes in rosacea. Our previous bulk RNA sequencing study on rosacea whole and epidermal skin also showed that aberrant activation of epidermal IFN/STAT1 is shared across
ETR, PPR and PHR Subtypes34. To be mentioned, although type I IFNs are not detected, our results here show that type I IFN signaling is among the top upregulated pathways in rosacea. We
speculate that type I and II IFN signaling pathways share very similar downstream genes; although these shared genes are upregulated in response to type II rather than type I IFN, they were
also included in the gene set of type I IFN signaling pathway when GSEA was performed. Collectively, these findings might provide an upstream explanation for the impaired skin barrier in
rosacea. However, a recent study has shown that the expression of type I IFNs, including _IFNA2_ and _IFNB1_, is significantly increased in plasmacytoid dendritic cells of the lesional skin
from rosacea patients during acute flares, but not changed during stable disease. They further revealed dysbiotic commensal bacteria are responsible for the production of type I IFNs. In
addition, blockade of type I IFN signaling could partially improve the rosacea-like phenotypes in the same rosacea mouse model used in our study56, but the treatment appeared to be less
effective than blockade of IFNγ. Therefore, it is assumed that type I IFNs might only be produced in skin lesions with dysbiotic commensal bacteria, which may be an aggravating factor rather
than an initiating factor for rosacea development. Moreover, the products of microbiome can be recognized by keratinocytes and cells of the innate immune system, then activate Toll-like
receptors; further activation of inflammatory receptors can aggravate the impaired skin barrier, which may result in more inflammation6,56,57,58. Here, our results show that antigen
processing and presentation and Toll-like receptor signaling pathways are upregulated in multiple cell types, including keratinocytes, macrophages and Schwann cells, suggesting a possible
link between abnormal microbiome and inflammation. However, further study is needed to clarify the direct role of microbiome in rosacea. Increasing evidence has demonstrated that rosacea is
a kind of neurogenic skin inflammation5,6,38,39. Our recent study also identified multiple genetic variants associated with neurogenic inflammation, which can promote rosacea development by
modulating the production of neuropeptides in peripheral neurons36. However, the cellular and molecular changes in the local cutaneous neural system remain largely unclear. In the present
study, although neuron cells are not found in our scRNA-seq datasets, possibly due to the fact that the cell bodies of neurons are not located in skin, we uncover a cluster of Schwann cells,
a type of glial cell that surrounds the neurons and plays a critical role in the maintenance and function of peripheral nerves59. Previous studies have revealed neuroinflammatory roles of
Schwann cells in multiple inflammatory neuropathies, such as Chronic inflammatory demyelinating polyneuropathy (CIDP) and Guillain-Barre syndrome (GBS)60,61,62,63. Consistently, we here
identify a previously undefined subpopulation of Schwann cells unique to rosacea skin lesions, which is characterized by the upregulation of multiple pro-inflammatory pathways. However,
further study is needed to illustrate the precise mechanisms by which Schwann cells participate in the pathogenesis of rosacea, and it will be very interesting to figure out the landscape of
peripheral neural signatures and their interaction with other cells in the skin lesions of rosacea possibly by burgeoning technologies, such as single-cell spatial proteomics. Rosacea is
well established as a chronic inflammatory skin disorder, and the dominating infiltrating cells are T cells, macrophages and mast cells9. Therein, Th1/Th17 polarization and macrophage
infiltration are considered as an undervalued hallmark across all subtypes of rosacea64; mast cell activation has also been suggested to be involved in skin inflammation of rosacea65,66. We
here elucidate the landscape of cutaneous immune cells at single-cell level in the lesional skin of different subtypes of rosacea. In agreement with previous studies, T cells are the major
infiltrating cells, and Th1/Th17 cell differentiation is significantly increased across all rosacea subtypes, especially in PPR. Unexpectedly, we find that resident memory T cells are
expanded in rosacea, which might provide a cellular explanation for the easy recurrence of this disorder. Macrophages, considered as the master regulators of inflammation67, are increased in
all subtypes of rosacea, and express high levels of multiple T cell-recruiting chemokines, such as CCL5, CXCL9, CXCL10, CXCL11. Moreover, B cell infiltration is shown to be increased in PPR
skin lesions, but the potential roles need further study to clarify. In addition to inflammation, vascular dysfunction, including validation and angiogenesis, is another hallmark of
rosacea6,9,11,38. We here demonstrate that the percentage of vascular endothelial cells is increased in skin lesions of rosacea, especially in PPR and PhR, which further supports our
previous findings showing that angiogenesis is augmented and might be necessary for rosacea development68. Besides endothelial cells, blood vessels contain a cluster of vascular mural cells
(including vSMCs and pericytes) that make up most of the vessel walls, which are essential for vascular contraction and relaxation69. Here, though changes in muscle contraction are not
identified in our scRNA-seq datasets, we show that the phosphorylation levels of MLC2 are greatly decreased in vMCs in all subtypes of rosacea, even in the non-lesional skin, which might be
the direct cause of abnormal validation of blood vessels in rosacea. We speculate that the muscle contraction of vMCs is likely determined by post-transcriptional regulation, such as protein
phosphorylation, in rosacea. Various studies have roughly revealed the cellular and molecular alterations through global gene analysis and immunohistochemistry7,12,13,14,15,16, and our
present study illustrates the changes of different cell types in rosacea skin lesions at single-cell level, but the intercellular communications among cell types remain largely unknown. In
the present study, via cell-cell communication analysis, we identify fibroblasts as the leading cell type producing outgoing signals, and T cells, macrophages, vMCs, keratinocytes and
endothelial cells are the main target cells for these outgoing signals, suggesting a pivotal role for fibroblasts in the pathogenesis of rosacea. Previous scRNA-seq studies showed that
fibroblasts could contribute to the amplification of inflammatory responses via transition to a pro-inflammatory state and production of multiple chemokines, both in psoriasis and atopic
dermatitis. For instance, these fibroblasts could produce CCL19, connected by ligand-receptor interactions to CCR7+ dendritic cells in the lesional skin21,70. Consistently, we here
demonstrate that in rosacea, fibroblasts are reprogrammed into a pro-inflammatory state, featured by expressing high levels of disease-characteristic chemokines, like CCL19, CXCL1, CXCL2 and
CXCL12. Specifically, these reprogrammed fibroblasts generate abundant CCL19, which recruits CCR7+ T cells, the dominant infiltrating cells, by ligand-receptor interactions in the lesional
skin of rosacea. In addition to the pro-inflammatory outgoing signals, fibroblasts are also identified as the leading cell type producing outgoing signals related to vascular relaxation.
Among these vascular relaxation-associated signals, PTGDS, an enzyme that catalyzes the conversion of prostaglandin H2 (PGH2) to prostaglandin D2 (PGD2), is specifically expressed and
remarkably upregulated in fibroblasts of the lesional skin of all rosacea subtypes. As a consequence, PGD2, a classical mediator of vasodilation53,54, is significantly increased in the skin
lesions. As expected, PTGDR, the receptor of PGD2, is found to be specifically and highly expressed in the cutaneous vMCs, suggesting that fibroblasts contribute to abnormal vasodilation via
PGD2-PTGDR axis in rosacea. We conclude that fibroblasts are the leading cell type producing pro-inflammatory and vasodilative signals in the pathogenesis of rosacea. Our study is not
without limitations. First, due to the small sample size used in this study, further investigations are needed to elucidate some assumptions directly drawn from our scRNA-seq datasets.
Second, considering that only female human samples are used, we can not rule out that all of the present findings apply to the male, and further experiments will be needed to clarify this
issue. Third, given that the healthy skin samples were dominated by keratinocytes, this caveat should be kept in mind regarding the findings for cellular proportion, especially for the cell
type in which less than 10 cells are collected. In summary, the present study illustrates the cellular and molecular landscape of skin-resident cell populations, and determines fibroblasts
as the pivotal determinant in the development of rosacea, emphasizing the role of mesenchymal subpopulations in the treatment of this disorder. METHODS HUMAN SKIN SAMPLES This study was
approved by the ethical committee of the Xiangya Hospital of Central South University (No. 201404361). All skin biopsies were collected from the central facial skin of female healthy
volunteers, and lesional skin of the central face and corresponding normal skin surrounding the auricle from female patients with ETR, PPR and PhR (aged 20–45 years) from the Dermatology of
Xiangya Hospital, Central South University. Patients were diagnosed with rosacea by clinical and pathologic examination. The use of human samples was reviewed and approved in advance by the
Ethical Committee of the Xiangya Hospital, Central South University, and written informed consent was obtained from all participants, and the participants received no compensation for their
participation. All experiments were conducted according to the principles set out in the WMA Declaration of Helsinki and the Department of Health and Human Services Belmont Report.
PREPARATION OF SINGLE-CELL SUSPENSION OF HUMAN SKIN BIOPSIES The whole tissue processing and the single-cell preparation process were completed within 3 h after the collection of surgical
skin biopsies. After removal of subcutaneous fat, skin samples were placed in a dish with 5 ml of dispase solution by dermis side down and incubated at 37 °C with shaking (80 rpm) for 60
min. Epidermis was carefully isolated from the dermis with forceps. The separated epidermis was placed in 5 ml of trypsin at 37 °C for 10 min, and the separated dermis was digested with 10
ml of collagenase V for 45 min, then neutralized with 5% FBS in PBS. The single-cell suspension was acquired by repeatedly aspirating and dispensing the solution with a 1-ml pipette and then
filtered through 70 μm and 40 μm strainers. The dead cells were removed with Dead Cell Removal Kit (Miltenyi Biotec). For each sample, all of the epidermis and dermis cells were pooled
together for the next step of single-cell analysis. SCRNA-SEQ scRNA-seq primarily involves GEM (gel bead-in-emulsion) generation, barcoding, cDNA amplification, library construction and
sequencing. These steps were performed in accordance with the user’s instructions of Chromium Single Cell 3ʹ Reagent Kits v3.1 (10× Genomics). Libraries were sequenced by an Illumina
NovaSeq6000 System. Approximately 10,000 cells (targeting 5000–12,000) per sample were subjected to single-cell RNA sequencing. PROCESSING OF SCRNA-SEQ DATA The raw reads data were mapped to
the hg38 human genome to perform quality control and the read counting of Ensemble genes using Cell Ranger (v6.0.2) (http://10xgenomics.com) with default parameters. Seurat (v4.3.0) R
package was used to exclude cells with fewer than 200 genes or more than 6000 genes detected and more than 20% mitochondrial reads for downstream analysis71. After quality control, 131,243
cells remained and were used for downstream bioinformatic analysis. The “SCTransform” function with default parameters was used to normalize and scale the feature expression measurements for
each cell by the total expression. The initial cell clustering was performed by “FindClusters” function by their first 20 PCs with Louvain algorithm at a resolution of 0.6. Non-linear
dimensional reduction was performed by “RunUMAP” function and visualized by Uniform Manifold Approximation and Projection (UMAP). Markers genes of each cell cluster were determined by
“FindAllMarkers” function with Wilcoxon rank-sum test. Only those with |‘avg_logFC’| ≥0.25 and ‘p_val_adj’ ≤0.05 were considered as marker genes. For subclustering analysis of each major
cell type, cells were extracted separately and clustered by their first 20 PCs and appropriate resolution. Markers genes of each subcluster were identified by “FindAllMarkers” function with
the default parameters. “DoHeatmap” function was used to show the top 20 markers gene in heatmap. PROFILING DIFFERENTIALLY EXPRESSED GENES WITHIN EACH CONDITION To identify differentially
expressed genes between lesional and non-lesional skin, we applied the “FindMarkers” function from Seurat with the following parameters: min.pct >0.25 and thresh.use = 0.25. The adjusted
_p_-value of each differentially expressed gene was calculated by non-parametric two-sided Wilcoxon rank-sum test and only those with |‘avg_logFC’| >0.25 and ‘p_val_adj’ < 0.05 were
considered to be differentially expressed genes. The gene expression levels shown in the various charts in the manuscript were plotted using packages in R. FUNCTION ENRICHMENT ANALYSIS Gene
Set Enrichment Analysis (GSEA) of GO and KEGG was performed by clusterProfiler R package (v4.5.1.902) and visualized with ggplot2 R package (v3.4.1)72. Representative terms that were
significantly enriched in at least one of the three rosacea subtypes, or top-ranked GO terms and KEGG pathways from MsigDB, were displayed. CELL-CELL COMMUNICATION ANALYSIS To assess
cell-cell communications among different cell types, we used CellChat (v1.1.3) to infer the intercellular communication network from single-cell RNA-seq data. Only cell types with more than
10 cells were considered in this analysis. The “trimean” method is used for calculating the average gene expression in “computeCommunProb” function. Pairwise comparison and visualization are
performed by CellChat build-in function, only interactions with _P_-value lower than 0.05 are considered to be significant. MOUSE MODEL OF ROSACEA AND TREATMENTS BALB/c and C57BL/6J
wildtype mice were purchased from Slark Company. The dorsal skin of the mice, aged 7 to 8 weeks, was shaved 1 day before the beginning of treatments. For the mouse model of rosacea,
intradermal injection of 40 μl LL37 (320 μM, Sangon Biotech) was applied on the dorsal skin twice a day for 2 days, as previously described10,16,36. For IFNγ blocking, neutralizing IFNγ
antibodies (10 μg/mice; eBioscience) were subcutaneously injected daily before and during the LL37 injection for a total of 3 days in BALB/c wildtype mice aged 8 weeks. The first treatment
dose began one day prior to the intradermal injection of LL37. An identical dose of IgG was administered to the control mouse. To deplete local skin fibroblasts, _Col1a2-_CreER mice with
C57BL/6J background (Jackson Laboratory, Strain #029567) were mated with iDTR mice with C57BL/6J background (Jackson Laboratory, Strain #007900). _Col1a2-_CreER/iDTR double positive
(Col1a2DTR) and WT mice were treated with tamoxifen (150 mg/kg, Sigma) daily for continuous five days at 5 weeks of age by intraperitoneal injection. 1 week after tamoxifen treatment, mice
were intradermally injected with diphtheria toxin (DT, 100 ng per mice, sigma) once daily for 3 days. The identical location that had received DT was intradermally injected with LL37 one
week later. For siRNA-mediated Ptgds knockdown in mice, _Ptgds_ or scrambled siRNAs (20 μL of 20 μM, purchased from the GenePharma company) were intradermally injected at the indicated site
of dorsal skin at indicated time in BALB/c wildtype mice aged 7 weeks. The oligo sequences of siRNAs used in this study are provided in Supplementary Data 11. The dorsal skin lesions were
observed using a stereoscope and harvested for subsequent experiments (including histological analysis, immunohistochemistry, immunofluorescence, ELISA, RT-qPCR, etc.) 48 h after the first
LL37 injection in all mice studies, and were scored for redness and area as previously described36. All mouse experiments were repeated three times, and 5–6 mice were included in each group
in each experiment. All mouse experiments were performed on both female and male mice, and there is no sex difference in the above mouse experiments, and the results of female mice were
presented in this study. Mice were randomly assigned to different groups and sex-matched in each experiment. All mice used in the presented study were housed in specific pathogen-free
conditions with a regular 12 h light/12 dark cycle at ~20–25 °C and 45–55% humidity and were fed standard rodent chow and tap water. Animal welfare was monitored, euthanasia was conducted
according to the guidelines, and the ethical committee of the Xiangya Hospital of Central South University approved all animal procedures. HISTOLOGICAL ANALYSIS The dorsal skin tissues of
the mice were fixed and embedded in paraffin. Paraffin sections were deparaffinized and stained with H&E solution. To examine histological changes, we count the number of infiltrating
cells in the dermis. Five randomly selected locations in each section were used to calculate the number of infiltrating cells in the dermis. IMMUNOHISTOCHEMISTRY Immunohistochemistry for
mouse skin sections (5 μm) was conducted as previously described36. To assess the vasodilation of cutaneous blood vessels, the perimeter of CD31-positive vessels was measured by ImageJ.
Double immunohistochemistry of human or mouse skin sections was performed using Opal™ 7-Color Manual IHC Kit according to the manufacturer’s protocols. Briefly, slides were deparaffinization
and rehydration. Before epitope retrieval, slides were fixed in 10% neutral buffered formalin for 30 min. Slides were cooled down at room temperature and washed twice with TBST (TBS buffer
containing 0.05% TritonX-100). Slides were covered with blocking buffer for 10 min, followed by incubation with primary antibody overnight at 4 °C. Slides were washed and incubated with
Polymer HRP secondary antibody for 10 min at room temperature. Slides were then washed and incubated with appropriate Opal Fluorophore to amplify the fluorescence signal. Multiplex staining
was performed by stripping the previous antibody in AR buffer before blocking and probing with the next primary antibody. Following similar steps from the first round of staining, slides
were developed to stain for other targets. Slides were counter-stained with DAPI for 5 min, and coverslipped with mounting medium. For CD31 staining, DAB staining kit (PV-9001, ZSGB-BIO) was
used according to the manufacturer’s instructions. The fluorescence intensity was assessed with ImageJ. The following primary antibodies were used: Rabbit anti-CD31 (1:100, 77699, Cell
Signaling), Mouse anti-KRT14 (1:1000, ab7800, abcam), Rabbit anti-KRT14 (1:2000, ab181595, abcam), Rabbit anti-CD74 (1:500, 77274, Cell signaling), Rabbit anti-CD74 (1:2000, ab289885,
abcam), Rabbit anti-CLDN4 (1:2000, ab210796, abcam), Rabbit anti-complement C3 (1:400, HPA003563, Atlas Antibodies), Rabbit anti-DCN (1:2000, ab277636, abcam), Rabbit anti-MBP (1:1500,
78896, Cell signaling), Rabbit anti-CD69 (1:500, ab233396, abcam), Rabbit anti-CD103 (1:2000, ab224202, abcam), Rabbit anti-p-MLC2 (1:200, 3671, Cell signaling), Mouse anti-α-SMA (1:5000,
ab7817, abcam), Mouse anti-CCL19 (1:100, MAB361, R&D systems), Mouse anti-CD4 (1:200, 14-2444-80, Thermo), Mouse anti-CCR7 (1:100, MAB197, R&D systems), Rabbit anti-PTGDS (1:200,
HPA004938, Atlas Antibodies), Rabbit anti-Vimentin (1:400, 5741, Cell signaling), Rabbit anti-PTGDR (1:200, HPA049668, Atlas Antibodies). Rabbit anti-p-STAT1 (1:1000, 9167, Cell signaling),
Rabbit anti-IRF1 (1:200, 8478, Cell signaling). IMMUNOFLUORESCENCE Skin lesions from human or mice were obtained and embedded in OCT. The sections were fixed with 4% paraformaldehyde and
blocked with 5% donkey serum in PBS for 1 h at room temperature. The sections were incubated with primary antibodies overnight (4 °C) and then were incubated with secondary antibodies for 1
h (room temperature). DAPI staining was used to indicate the cell nucleus. The quantification of fluorescence intensity was conducted with ImageJ, as previously described36. The following
primary antibodies were used: Goat anti-PDGFRA antibody (1:100, AF1062, R&D systems), Rabbit anti-PTGDS antibody (1:200, 10004344, Cayman), Rabbit anti-PDGFRA (1:500, ab203491, abcam),
Goat anti-CCL19 (1:100, AF880, R&D systems), Rabbit anti-CD4 (1:500, ab183685, abcam), Rat anti-CCR7 (1:100, MAB3477, R&D systems), Rat anti-IFNγ (1:100, 14-7311-81, Thermo), Rabbit
anti-MBP (1:1500, 78896, Cell signaling), Sheep anti-CD74 (1:100, AF7478, R&D systems), Rat anti-F4/80 (1:100, 14-4801-81, Thermo), Goat anti-CXCL10 antibody (1:100, AF466, R&D
systems), Rabbit anti-MLC2 (1:400, 15354-1-AP, Proteintech), Mouse anti-α-SMA (1:5000, ab7817, abcam). MEASUREMENT OF TEWL TEWL was assessed with Tewameter® TM 300 (Courage + Khazaka
Electronic, Germany). An average value of three repeated measurements was used for each mouse, and all measurements were conducted by the same investigator. ENZYME-LINKED IMMUNOSORBENT ASSAY
(ELISA) The concentration of Prostaglandin D2 (PGD2) in skin lesions from rosacea patients, healthy individuals and mice was quantified using a PGD2 ELISA kit (512031-96stripwells, Cayman)
according to the manufacturer’s protocol. CELL CULTURE AND TREATMENT Primary human keratinocytes were isolated from human foreskin (aged 2–5) and cultured in CnT-07 (CELLnTEC, USA). For IFNγ
treatment, at a confluency of 40%, cells were treated with IFNγ (500 IU/ml) for 48 h, then the cells were subjected to subsequent analysis. RT-QPCR Total RNA from skin lesion and
keratinocytes was extracted by TRIzol reagent (Thermo) and then reverse-transcribed to cDNA using the PrimeScript RT Reagent Kit (Thermo). qPCR was performed using 2x Taq Pro Universal SYBR
qPCR Master Mix (Vazyme). The relative mRNA expression levels were calculated based on the 2−ΔΔCT method. The sequences of the primers used in this study are provided in Supplementary Data
11. STATISTICAL ANALYSIS Statistical analyses were performed using GraphPad Prism (version 8). All data are presented as the mean ± SEM. We determined the data for normal distribution and
similar variance between groups. The differences between 2 groups were compared by 2-tailed unpaired or paired Student’s _t_-test. For comparisons of more than 2 groups, one-way ANOVA with
Tukey’s post hoc test or two-way ANOVA with a post hoc Holm–Sidak’s multiple comparisons test was performed. Two-sided permutation test without multiple comparison adjustments was used for
GSEA analysis. When the data were not normally distributed or exhibited unequal variances between groups, we performed statistical analysis with two-tailed Mann–Whitney _U_ test. No
statistical method was employed to predetermine the sample size. Mice in this study were randomly allocated to different groups. REPORTING SUMMARY Further information on research design is
available in the Nature Portfolio Reporting Summary linked to this article. DATA AVAILABILITY All data needed to evaluate the conclusions in this study are provided in the manuscript and/or
the Supplementary Materials. The scRNA-seq datasets are available from the genome sequence archive under accession number HRA006167 (http://bigd.big.ac.cn/gsa-human/). Bulk RNA-seq datasets
for the skin lesions of rosacea patients used in this study were obtained from the genome sequence archive under accession number HRA000379 (http://bigd.big.ac.cn/gsa-human/). All relevant
approvals were obtained from China’s Ministry of Science and Technology related to the export of genetic information and materials relevant to this work. Any other details supporting the
findings of the present study are available from the corresponding author upon reasonable request. Source data are provided with this paper. REFERENCES * van Zuuren, E. J. Rosacea. _N. Engl.
J. Med._ 377, 1754–1764 (2017). Article PubMed Google Scholar * Gether, L., Overgaard, L. K., Egeberg, A. & Thyssen, J. P. Incidence and prevalence of rosacea: a systematic review
and meta-analysis. _Br. J. Dermatol._ 179, 282–289 (2018). CAS PubMed Google Scholar * Li, J. et al. Epidemiological features of rosacea in Changsha, China: a population-based,
cross-sectional study. _J. Dermatol._ 47, 497–502 (2020). Article PubMed Google Scholar * Wilkin, J. et al. Standard classification of rosacea: report of the National Rosacea Society
Expert Committee on the classification and staging of rosacea. _J. Am. Acad. Dermatol._ 46, 584–587 (2002). Article PubMed Google Scholar * Gallo, R. L. et al. Standard classification and
pathophysiology of rosacea: the 2017 update by the National Rosacea Society Expert Committee. _J. Am. Acad. Dermatol._ 78, 148–155 (2018). Article PubMed Google Scholar * Buddenkotte, J.
& Steinhoff, M. Recent advances in understanding and managing rosacea. _F1000Res_ 7, F1000 Faculty Rev-1885 (2018). Article PubMed Google Scholar * Medgyesi, B. et al. Rosacea is
characterized by a profoundly diminished skin barrier. _J. Invest. Dermatol._ 140, 1938–1950.e1935 (2020). Article CAS PubMed Google Scholar * Deng, Z. et al. Claudin reduction may
relate to an impaired skin barrier in rosacea. _J. Dermatol._ 46, 314–321 (2019). Article CAS PubMed Google Scholar * Steinhoff, M. et al. Clinical, cellular, and molecular aspects in
the pathophysiology of rosacea. _J. Investig. Dermatol. Symp. Proc._ 15, 2–11 (2011). Article CAS PubMed PubMed Central Google Scholar * Deng, Z. et al. A positive feedback loop between
mTORC1 and cathelicidin promotes skin inflammation in rosacea. _EMBO Mol. Med._ 13, e13560 (2021). Article CAS PubMed PubMed Central Google Scholar * Liu, T. et al. Aberrant amino acid
metabolism promotes neurovascular reactivity in rosacea. _JCI Insight_ 7, e161870 (2022). Article PubMed PubMed Central Google Scholar * Zhang, H. et al. Murine models of rosacea: a
review. _J. Cosmet. Dermatol._ 21, 905–909 (2022). Article PubMed Google Scholar * Ahn, C. S. & Huang, W. W. Rosacea pathogenesis. _Dermatol. Clin._ 36, 81–86 (2018). Article CAS
PubMed Google Scholar * Shih, Y. H., Xu, J., Kumar, A., Li, R. & Chang, A. L. S. Alterations of immune and keratinization gene expression in papulopustular rosacea by whole
transcriptome analysis. _J. Invest. Dermatol._ 140, 1100–1103.e1104 (2020). Article CAS PubMed Google Scholar * Woo, Y. R., Lim, J. H., Cho, D. H. & Park, H. J. Rosacea: molecular
mechanisms and management of a chronic cutaneous inflammatory condition. _Int. J. Mol. Sci._ 17, 1562 (2016). Article PubMed PubMed Central Google Scholar * Yamasaki, K. et al. Increased
serine protease activity and cathelicidin promotes skin inflammation in rosacea. _Nat. Med._ 13, 975–980 (2007). Article CAS PubMed Google Scholar * Papalexi, E. & Satija, R.
Single-cell RNA sequencing to explore immune cell heterogeneity. _Nat. Rev. Immunol._ 18, 35–45 (2018). Article CAS PubMed Google Scholar * Potter, S. S. Single-cell RNA sequencing for
the study of development, physiology and disease. _Nat. Rev. Nephrol._ 14, 479–492 (2018). Article CAS PubMed PubMed Central Google Scholar * Mereu, E. et al. Benchmarking single-cell
RNA-sequencing protocols for cell atlas projects. _Nat. Biotechnol._ 38, 747–755 (2020). Article CAS PubMed Google Scholar * Kim, D., Chung, K. B. & Kim, T. G. Application of
single-cell RNA sequencing on human skin: technical evolution and challenges. _J. Dermatol. Sci._ 99, 74–81 (2020). Article CAS PubMed Google Scholar * He, H. et al. Single-cell
transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. _J. Allergy Clin. Immunol._ 145, 1615–1628 (2020).
Article CAS PubMed Google Scholar * Liu, X. et al. Single-cell RNA-sequencing reveals lineage-specific regulatory changes of fibroblasts and vascular endothelial cells in keloids. _J.
Invest. Dermatol._ 142, 124–135.e111 (2022). Article CAS PubMed Google Scholar * Hughes, T. K. et al. Second-strand synthesis-based massively parallel scRNA-Seq reveals cellular states
and molecular features of human inflammatory skin pathologies. _Immunity_ 53, 878–894.e877 (2020). Article CAS PubMed PubMed Central Google Scholar * Sole-Boldo, L. et al. Single-cell
transcriptomes of the human skin reveal age-related loss of fibroblast priming. _Commun. Biol._ 3, 188 (2020). Article CAS PubMed PubMed Central Google Scholar * Zheng, M. et al.
Single-cell sequencing shows cellular heterogeneity of cutaneous lesions in lupus erythematosus. _Nat. Commun._ 13, 7489 (2022). Article ADS CAS PubMed PubMed Central Google Scholar *
Jiang, Y. et al. Cytokinocytes: the diverse contribution of keratinocytes to immune responses in skin. _JCI Insight_ 5, e142067 (2020). Article PubMed PubMed Central Google Scholar *
Kabashima, K., Honda, T., Ginhoux, F. & Egawa, G. The immunological anatomy of the skin. _Nat. Rev. Immunol._ 19, 19–30 (2019). Article CAS PubMed Google Scholar * Ter Horst, B.,
Chouhan, G., Moiemen, N. S. & Grover, L. M. Advances in keratinocyte delivery in burn wound care. _Adv. Drug Deliv. Rev._ 123, 18–32 (2018). Article PubMed PubMed Central Google
Scholar * Kim, Y. M. & Shin, E. C. Type I and III interferon responses in SARS-CoV-2 infection. _Exp. Mol. Med._ 53, 750–760 (2021). Article CAS PubMed PubMed Central Google Scholar
* Kanoh, H. et al. IFN-gamma reduces epidermal barrier function by affecting fatty acid composition of ceramide in a mouse atopic dermatitis model. _J. Immunol. Res._ 2019, 3030268 (2019).
Article PubMed PubMed Central Google Scholar * Mizutani, Y., Takagi, N., Nagata, H. & Inoue, S. Interferon-gamma downregulates tight junction function, which is rescued by
interleukin-17A. _Exp. Dermatol._ 30, 1754–1763 (2021). Article CAS PubMed PubMed Central Google Scholar * Hile, G. A., Gudjonsson, J. E. & Kahlenberg, J. M. The influence of
interferon on healthy and diseased skin. _Cytokine_ 132, 154605 (2020). Article CAS PubMed Google Scholar * Srivastava, A. et al. Cross-talk between IFN-gamma and TWEAK through miR-149
amplifies skin inflammation in psoriasis. _J. Allergy Clin. Immunol._ 147, 2225–2235 (2021). Article CAS PubMed Google Scholar * Deng, Z. et al. Keratinocyte-immune cell crosstalk in a
STAT1-mediated pathway: novel insights into rosacea pathogenesis. _Front. Immunol._ 12, 674871 (2021). Article CAS PubMed PubMed Central Google Scholar * Wei, K., Nguyen, H. N. &
Brenner, M. B. Fibroblast pathology in inflammatory diseases. _J. Clin. Invest._ 131, e149538 (2021). Article CAS PubMed PubMed Central Google Scholar * Deng, Z. et al. Whole genome
sequencing identifies genetic variants associated with neurogenic inflammation in rosacea. _Nat. Commun._ 14, 3958 (2023). Article ADS CAS PubMed PubMed Central Google Scholar * Zhu,
Y., Duan, S., Wang, M., Deng, Z. & Li, J. Neuroimmune interaction: a widespread mutual regulation and the weapons for barrier organs. _Front. Cell Dev. Biol._ 10, 906755 (2022). Article
PubMed PubMed Central Google Scholar * Schwab, V. D. et al. Neurovascular and neuroimmune aspects in the pathophysiology of rosacea. _J. Investig. Dermatol. Symp. Proc._ 15, 53–62
(2011). Article CAS PubMed PubMed Central Google Scholar * Aubdool, A. A. & Brain, S. D. Neurovascular aspects of skin neurogenic inflammation. _J. Investig. Dermatol. Symp. Proc._
15, 33–39 (2011). Article CAS PubMed Google Scholar * Wang, S. et al. S100A8/A9 in Inflammation. _Front. Immunol._ 9, 1298 (2018). Article PubMed PubMed Central Google Scholar *
Zhang, J. Yin and yang interplay of IFN-gamma in inflammation and autoimmune disease. _J. Clin. Invest._ 117, 871–873 (2007). Article CAS PubMed PubMed Central Google Scholar *
Gauthier, M. et al. CCL5 is a potential bridge between type 1 and type 2 inflammation in asthma. _J. Allergy Clin. Immunol._ 152, 94–106.e112 (2023). Article CAS PubMed PubMed Central
Google Scholar * Cano-Gamez, E. et al. Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4+ T cells to cytokines. _Nat. Commun._ 11, 1801 (2020).
Article ADS CAS PubMed PubMed Central Google Scholar * Salkowska, A. et al. Identification of novel molecular markers of human Th17 cells. _Cells_ 9, 1611 (2020). Article CAS PubMed
PubMed Central Google Scholar * Bao, K. & Reinhardt, R. L. The differential expression of IL-4 and IL-13 and its impact on type-2 immunity. _Cytokine_ 75, 25–37 (2015). Article CAS
PubMed PubMed Central Google Scholar * Zhang, J. Y. et al. Single-cell landscape of immunological responses in patients with COVID-19. _Nat. Immunol._ 21, 1107–1118 (2020). Article CAS
PubMed Google Scholar * Siekmann, A. F. Biology of vascular mural cells. _Development_ 150, dev200271 (2023). Article CAS PubMed PubMed Central Google Scholar * Yemisci, M. et al.
Pericyte contraction induced by oxidative-nitrative stress impairs capillary reflow despite successful opening of an occluded cerebral artery. _Nat. Med._ 15, 1031–1037 (2009). Article CAS
PubMed Google Scholar * Gonzales, A. L. et al. Contractile pericytes determine the direction of blood flow at capillary junctions. _Proc. Natl Acad. Sci. USA_ 117, 27022–27033 (2020).
Article ADS CAS PubMed PubMed Central Google Scholar * Korte, N. et al. The Ca2+-gated channel TMEM16A amplifies capillary pericyte contraction and reduces cerebral blood flow after
ischemia. _J. Clin. Invest._ 132, e154118 (2022). Article CAS PubMed PubMed Central Google Scholar * Nayak, A. et al. Single-molecule analysis reveals that regulatory light chains
fine-tune skeletal myosin II function. _J. Biol. Chem._ 295, 7046–7059 (2020). Article CAS PubMed PubMed Central Google Scholar * Jin, S. et al. Inference and analysis of cell-cell
communication using CellChat. _Nat. Commun._ 12, 1088 (2021). Article ADS CAS PubMed PubMed Central Google Scholar * Morrow, J. D., Parsons, W. G. 3rd & Roberts, L. J. Release of
markedly increased quantities of prostaglandin D2 in vivo in humans following the administration of nicotinic acid. _Prostaglandins_ 38, 263–274 (1989). Article CAS PubMed Google Scholar
* Kong, D. & Yu, Y. Prostaglandin D2 signaling and cardiovascular homeostasis. _J. Mol. Cell Cardiol._ 167, 97–105 (2022). Article CAS PubMed Google Scholar * Addor, F. A. Skin
barrier in rosacea. _Bras. Dermatol._ 91, 59–63 (2016). Article Google Scholar * Mylonas, A. et al. Type I IFNs link skin-associated dysbiotic commensal bacteria to pathogenic inflammation
and angiogenesis in rosacea. _JCI Insight_ 8, e151846 (2023). Article PubMed PubMed Central Google Scholar * Holmes, A. D. Potential role of microorganisms in the pathogenesis of
rosacea. _J. Am. Acad. Dermatol._ 69, 1025–1032 (2013). Article PubMed Google Scholar * Moran, E. M., Foley, R. & Powell, F. C. Demodex and rosacea revisited. _Clin. Dermatol._ 35,
195–200 (2017). Article PubMed Google Scholar * Bosch-Queralt, M., Fledrich, R. & Stassart, R. M. Schwann cell functions in peripheral nerve development and repair. _Neurobiol. Dis._
176, 105952 (2023). Article CAS PubMed Google Scholar * Ydens, E. et al. The neuroinflammatory role of Schwann cells in disease. _Neurobiol. Dis._ 55, 95–103 (2013). Article CAS PubMed
Google Scholar * Allard, D. E. et al. Schwann cell-derived periostin promotes autoimmune peripheral polyneuropathy via macrophage recruitment. _J. Clin. Invest._ 128, 4727–4741 (2018).
Article PubMed PubMed Central Google Scholar * Zhang, H. et al. Activated Schwann cells and increased inflammatory cytokines IL-1beta, IL-6, and TNF-alpha in patients’ sural nerve are
lack of tight relationship with specific sensory disturbances in Parkinson’s disease. _CNS Neurosci. Ther._ 26, 518–526 (2020). Article ADS CAS PubMed Google Scholar * Trias, E. et al.
Schwann cells orchestrate peripheral nerve inflammation through the expression of CSF1, IL-34, and SCF in amyotrophic lateral sclerosis. _Glia_ 68, 1165–1181 (2020). Article PubMed Google
Scholar * Buhl, T. et al. Molecular and morphological characterization of inflammatory infiltrate in rosacea reveals activation of Th1/Th17 pathways. _J. Invest. Dermatol._ 135, 2198–2208
(2015). Article CAS PubMed Google Scholar * Muto, Y. et al. Mast cells are key mediators of cathelicidin-initiated skin inflammation in rosacea. _J. Invest. Dermatol._ 134, 2728–2736
(2014). Article CAS PubMed PubMed Central Google Scholar * Mascarenhas, N. L., Wang, Z., Chang, Y. L. & Di Nardo, A. TRPV4 mediates mast cell activation in cathelicidin-induced
rosacea inflammation. _J. Invest. Dermatol._ 137, 972–975 (2017). Article CAS PubMed Google Scholar * Wynn, T. A. & Barron, L. Macrophages: master regulators of inflammation and
fibrosis. _Semin. Liver Dis._ 30, 245–257 (2010). Article CAS PubMed PubMed Central Google Scholar * Peng, Q. et al. mTORC1-mediated angiogenesis is required for the development of
rosacea. _Front. Cell Dev. Biol._ 9, 751785 (2021). Article PubMed PubMed Central Google Scholar * Basatemur, G. L., Jorgensen, H. F., Clarke, M. C. H., Bennett, M. R. & Mallat, Z.
Vascular smooth muscle cells in atherosclerosis. _Nat. Rev. Cardiol._ 16, 727–744 (2019). Article PubMed Google Scholar * Ma, F. et al. Single cell and spatial sequencing define processes
by which keratinocytes and fibroblasts amplify inflammatory responses in psoriasis. _Nat. Commun._ 14, 3455 (2023). Article ADS CAS PubMed PubMed Central Google Scholar * Hao, Y. et
al. Integrated analysis of multimodal single-cell data. _Cell_ 184, 3573–3587.e3529 (2021). Article CAS PubMed PubMed Central Google Scholar * Wu, T. et al. clusterProfiler 4.0: a
universal enrichment tool for interpreting omics data. _Innovation_ 2, 100141 (2021). CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS This work was
supported by the National Natural Science Funds for Distinguished Young Scholars (No. 82225039), the National Natural Science Funds for Excellent Young Scientists (No. 82422063), the
National Key Research and Development Program of China (No. 2023YFC2509003), the National Natural Science Foundation of China (No. 82373508, No. 82303992, No. 82203958, No. 82073457,
No.82203945, No. 82173448, No. 81874251), the Natural Science Funds of Hunan province for Excellent Young Scholars (No. 2023JJ20094), the Natural Science Foundation of Hunan Province, China
(No. 2021JJ31079), Educational Science Planning Project of Hunan Province (No. ND206997), and a Major Research Plan of the National Natural Science Foundation of China (No. 92374108). We
thank our colleagues (Department of Dermatology, Xiangya Hospital, Central South University, China) for their generous support throughout this work. AUTHOR INFORMATION Author notes * These
authors contributed equally: Mengting Chen, Li Yang. AUTHORS AND AFFILIATIONS * Department of Dermatology, Xiangya Hospital, Central South University, Changsha, Hunan, China Mengting Chen,
Li Yang, Zheng Wu, Zixin Tan, Wenqin Xiao, San Xu, Yan Zhu, Mei Wang, Dan Jian, Fangfen Liu, Yan Tang, Zhixiang Zhao, Yingxue Huang, Wei Shi, Hongfu Xie, Ben Wang, Zhili Deng & Ji Li *
Hunan Key Laboratory of Aging Biology, Xiangya Hospital, Central South University, Changsha, Hunan, China Mengting Chen, Li Yang, Zheng Wu, Zixin Tan, Wenqin Xiao, San Xu, Yan Zhu, Mei Wang,
Dan Jian, Fangfen Liu, Yan Tang, Zhixiang Zhao, Yingxue Huang, Wei Shi, Hongfu Xie, Ben Wang, Zhili Deng & Ji Li * National Clinical Research Center for Geriatric Disorders, Xiangya
Hospital, Central South University, Changsha, Hunan, China Mengting Chen, Li Yang, Zheng Wu, Zixin Tan, Wenqin Xiao, San Xu, Yan Zhu, Mei Wang, Dan Jian, Fangfen Liu, Yan Tang, Zhixiang
Zhao, Yingxue Huang, Wei Shi, Hongfu Xie, Ben Wang, Zhili Deng & Ji Li * FuRong Laboratory, Changsha, China Mengting Chen, Li Yang, Zheng Wu, Zixin Tan, Wenqin Xiao, San Xu, Yan Zhu, Mei
Wang, Dan Jian, Fangfen Liu, Yan Tang, Zhixiang Zhao, Yingxue Huang, Wei Shi, Hongfu Xie, Ben Wang, Zhili Deng & Ji Li * Center for Machine Learning Research, Peking University,
Beijing, China Peijie Zhou * AI for Science Institute, Beijing, China Peijie Zhou * School of Mathematics and Statistics, Wuhan University, Wuhan, China Suoqin Jin * Department of
Mathematics, University of California Irvine, Irvine, CA, USA Qing Nie * The NSF-Simons Center for Multiscale Cell Fate Research, University of California Irvine, Irvine, CA, USA Qing Nie *
Department of Developmental and Cell Biology, University of California Irvine, Irvine, CA, USA Qing Nie Authors * Mengting Chen View author publications You can also search for this author
inPubMed Google Scholar * Li Yang View author publications You can also search for this author inPubMed Google Scholar * Peijie Zhou View author publications You can also search for this
author inPubMed Google Scholar * Suoqin Jin View author publications You can also search for this author inPubMed Google Scholar * Zheng Wu View author publications You can also search for
this author inPubMed Google Scholar * Zixin Tan View author publications You can also search for this author inPubMed Google Scholar * Wenqin Xiao View author publications You can also
search for this author inPubMed Google Scholar * San Xu View author publications You can also search for this author inPubMed Google Scholar * Yan Zhu View author publications You can also
search for this author inPubMed Google Scholar * Mei Wang View author publications You can also search for this author inPubMed Google Scholar * Dan Jian View author publications You can
also search for this author inPubMed Google Scholar * Fangfen Liu View author publications You can also search for this author inPubMed Google Scholar * Yan Tang View author publications You
can also search for this author inPubMed Google Scholar * Zhixiang Zhao View author publications You can also search for this author inPubMed Google Scholar * Yingxue Huang View author
publications You can also search for this author inPubMed Google Scholar * Wei Shi View author publications You can also search for this author inPubMed Google Scholar * Hongfu Xie View
author publications You can also search for this author inPubMed Google Scholar * Qing Nie View author publications You can also search for this author inPubMed Google Scholar * Ben Wang
View author publications You can also search for this author inPubMed Google Scholar * Zhili Deng View author publications You can also search for this author inPubMed Google Scholar * Ji Li
View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS J.L., Z.D., M.C., B.W. and Q.N. designed and conceived the study. Z.D., J.L., L.Y. and
M.C. performed data analyses. L.Y., Z.D., P.Z. and S.J. performed scRNA-seq analysis. M.C. and Z.D. performed most experiments. Z.Z., W.X., W.S., D.J., B.W., F.L., Y.T., Y.H., Y.Z.
contributed to sample collection. Z.W., Z.T., S.X., Y.Z. and M.W. helped to perform mouse experiments. H.X. provided critical discussion and suggestions. Z.D., J.L., L.Y., M.C., B.W. and
Q.N. prepared the manuscript with input from coauthors. CORRESPONDING AUTHORS Correspondence to Qing Nie, Ben Wang, Zhili Deng or Ji Li. ETHICS DECLARATIONS COMPETING INTERESTS The authors
declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Communications_ thanks the anonymous reviewers for their contribution to the peer review of this work. A peer
review file is available. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION PEER REVIEW FILE DESCRIPTION OF ADDITIONAL SUPPLEMENTARY FILES SUPPLEMENTARY DATA 1 SUPPLEMENTARY DATA 2 SUPPLEMENTARY DATA 3
SUPPLEMENTARY DATA 4 SUPPLEMENTARY DATA 5 SUPPLEMENTARY DATA 6 SUPPLEMENTARY DATA 7 SUPPLEMENTARY DATA 8 SUPPLEMENTARY DATA 9 SUPPLEMENTARY DATA 10 SUPPLEMENTARY DATA 11 REPORTING SUMMARY
SOURCE DATA SOURCE DATA RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits
any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the
Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of
it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material
is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission
directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE
Chen, M., Yang, L., Zhou, P. _et al._ Single-cell transcriptomics reveals aberrant skin-resident cell populations and identifies fibroblasts as a determinant in rosacea. _Nat Commun_ 15,
8737 (2024). https://doi.org/10.1038/s41467-024-52946-7 Download citation * Received: 17 December 2023 * Accepted: 25 September 2024 * Published: 09 October 2024 * DOI:
https://doi.org/10.1038/s41467-024-52946-7 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative