Play all audios:
ABSTRACT The single-molecule tracking of transmembrane receptors in living cells has provided significant insights into signaling mechanisms, such as mobility and clustering upon their
activation/inactivation, making it a potential screening method for drug discovery. Here we show that single-molecule tracking-based screening can be used to explore compounds both
detectable and undetectable by conventional methods for disease-related receptors. Using an automated system for a fast large-scale single-molecule analysis, we screen for epidermal growth
factor receptor (EGFR) from 1134 of FDA approved drugs. The 18 hit compounds include all EGFR-targeted tyrosine kinase inhibitors (TKIs) in the library that suppress any
phosphorylation-dependent mobility shift of EGFR, proving the concept of this approach. The remaining hit compounds are not reported as EGFR-targeted drugs and do not inhibit EGF-induced
EGFR phosphorylation. These non-TKI compounds affect the mobility and/or clustering of EGFR without EGF and induce EGFR internalization, to impede EGFR-dependent cell growth. Thus,
single-molecule tracking provides an alternative modality for discovering therapeutics on various receptor functions with previously untargeted mechanisms. SIMILAR CONTENT BEING VIEWED BY
OTHERS TALES OF 1,008 SMALL MOLECULES: PHENOMIC PROFILING THROUGH LIVE-CELL IMAGING IN A PANEL OF REPORTER CELL LINES Article Open access 06 August 2020 MULTIPLEXED AND REPRODUCIBLE HIGH
CONTENT SCREENING OF LIVE AND FIXED CELLS USING DYE DROP Article Open access 14 November 2022 MULTIPLEXED PROFILING OF INTRACELLULAR PROTEIN ABUNDANCE, ACTIVITY, INTERACTIONS AND
DRUGGABILITY WITH LABEL-SEQ Article 21 October 2024 INTRODUCTION Transmembrane receptors trigger signal transduction to induce cellular responses. Since the downstream signaling depends on
the functional properties of receptors, various diseases are often attributed to receptor dysfunction. In drug discovery, the largest number of target molecules are membrane receptors
(>50% of total), such as receptor tyrosine kinases (RTKs), G protein-coupled receptors (GPCRs), and immune receptors, followed by nuclear receptors (<20%)1,2,3. Epidermal growth factor
receptor (EGFR), an RTK, mediates signal transduction for cell proliferation, differentiation, migration, and apoptosis, and is a primary target molecule in drug exploration because its
overexpression and/or mutations are found in various types of cancer cells4,5,6,7. Many small-molecule drugs against a well-known EGFR-related cancer, non-small cell lung cancer (NSCLC),
have been developed: gefitinib, erlotinib, and icotinib as first-generation tyrosine kinase inhibitors (TKIs); afatinib and dacomitinib as second-generation TKIs; and osimertinib as
third-generation8. These drugs have been evaluated by their inhibitory effects on tyrosine kinase activity9, but EGFR undergoes multiple signaling processes including tyrosine
phosphorylation, oligomerization, coupling with adapter molecules, and internalization on the membrane of living cells10. Disturbing these processes can affect the signaling activity of the
molecule, providing a potential mechanistic target for drug discovery. In general, assays for drug discovery focus on a particular step in the signaling pathway exhibited by the targeted
molecules. However, assays that select for compounds that have effects on multiple steps in the signaling pathway are of interest because of their different mechanisms of action on the same
target11. Single-molecule imaging analysis has been used to visualize and measure the location and brightness of individual membrane receptor molecules in cells12,13,14. These measurements
provide information on the lateral diffusion and oligomerization/clustering of receptors on the membrane of living cells. In the case of EGFR, the behavioral transition on the membrane,
which is described by EGFR mobility and clustering dynamics, has been shown to correlate to the signaling process15,16,17,18,19: EGF triggers tyrosine phosphorylation, leading to the
translocation of EGFR along the membrane to a specific region where the mobility of EGFR is confined and the clusters responsible for downstream signaling are formed, followed by
internalization into endosomes. Single-molecule imaging has also revealed other processes during EGFR signaling, such as ligand-receptor binding kinetics, molecular structure transitions
depending on membrane components, and colocalization with receptors for different signaling pathways. That is, single-molecule imaging has the potential to elucidate the effects of
pharmacological compounds on multiple events during a series of EGFR signaling processes. In particular, this technique can detect changes in the physical properties of the target receptor,
which include the lateral diffusion and the statistical distribution of receptor clusters, both of which are difficult to detect using biochemical methods. However, the low throughput of
single-molecule imaging for data acquisition, which arises from its expertize- and manual-dependent workflow, prevents this method from being employed for large-scale single-molecule
analysis. As a solution, we previously achieved a fully automated, in-cell single-molecule imaging system equipped with robotics and machine learning (AiSIS)20,21 that has a 100-fold higher
throughput than standard single-molecule analysis techniques. High-throughput analysis by AiSIS enables the evaluation of numerous compounds by detecting their effect on molecular mobility
and clustering during the signaling process, which provides a form of molecular-targeted drug screening. Here, we report a single-molecule tracking-based technique for the screening of
compounds targeting transmembrane receptors. The successful selection of all TKIs for EGFR from a library consisting of 1134 FDA-approved compounds provides the proof of concept of this
approach. The other hit compounds exhibit effects on the clustering, internalization, and expression level of EGFR without strong inhibition of the tyrosine kinase activity upon EGF
stimulation. Unlike screening methods that focus on a single event in the molecular process, our method can assess various physical events at the single-molecule level occurring during the
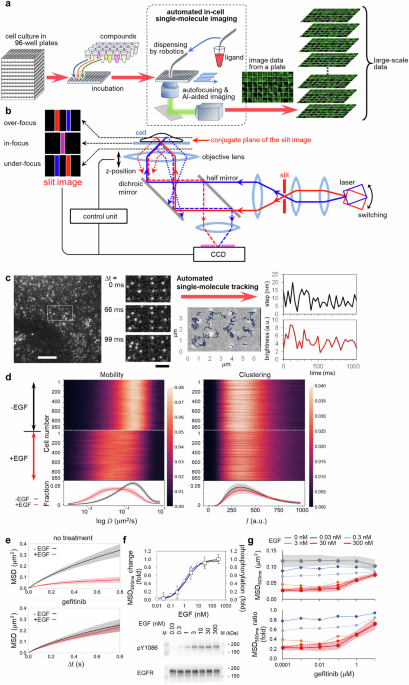
signal transduction of the target molecule. RESULTS EGFR MOBILITY-BASED SCREENING AiSIS allows for a large-scale, single-molecule analysis of plasma membrane receptors using 96-well plates
(Fig. 1a). Cells in each well were treated with a compound, and single-molecule imaging using total internal reflection fluorescence microscopy (TIRFM) was automatically executed well by
well. Autofocusing mechanism (Fig. 1b and Supplementary Fig. 1) kept the in-focus position precise to acquire high magnification images of living cells with clear single fluorescence spots
of the target molecules on the membrane by TIRFM. The fields of view suitable for the single-molecule analysis were determined by machine learning-based automated cell searching
(Supplementary Movie 1). During the imaging, autofocusing was continuously running, and the ligand was dispensed at desired times by robotics. In the case of measuring 20 cells under one
measurement condition, as described later, 480 different measurement conditions for ligand/compound concentrations, time of the addition, and duration of the treatment could be examined on
demand in a single assay in one day. By observing individual cells under uniform fluorescence excitation at high magnification, quantitative analysis of the position as well as the
fluorescence intensity of the fluorescent spots is possible, which allows the characterization of not only the lateral mobility of the target molecule but also the cluster formation of the
target molecule. Figure 1c and Supplementary Movie 2a show a single EGFR-mEGFP complex expressed in CHO-K1 cells, which lack endogenous EGFR. From the images acquired under optimized
conditions, trajectory data, including the positions and brightness of fluorescent EGFR, were obtained by single-molecule tracking (Fig. 1d) and used to quantify the mobility of EGFR with
diffusion coefficients and mean square displacements (MSD) (Fig. 1e). The mobility decreased with EGF addition, as shown by the MSD (Fig. 1f), consistent with a previous study20. This
EGF-induced mobility shift was recovered by treatment with a TKI (gefitinib; Fig. 1e, bottom, and Supplementary Movie 2b), which confirmed EGFR mobility tightly correlated with EGF-activated
kinase function. As shown in Fig. 1f, a linear relationship was found between the MSD for 500 ms (MSD500ms) and EGFR phosphorylation under various EGF concentrations. Both data were
obtained approximately 2 min after EGF addition, a time that represents the early phase of EGFR signaling just after reaching maximum EGFR phosphorylation. The inhibitory effect of gefitinib
on the EGF-induced decrease of MSD500ms is shown in Fig. 1g (top). Because TKI itself seems to have little effect on the mobility in our data, the MSD ratio (MSD with EGF to MSD without
EGF) was used to evaluate the compounds en bloc. As shown in Fig. 1g (bottom), when the EGF concentration was 30 nM, the ratio was approximately 0.2 for TKI-untreated cells but approached 1
for treated cells due to suppression of the EGF-induced decrease of EGFR mobility. We extended the single-molecule tracking-based assay to drug screening. First, we optimized the conditions
of the image acquisition (Supplementary Fig. 2), and the suitability of this approach as a screening method was assessed using the indices calculated from MSD500ms, with the positive control
being cells treated with gefitinib and EGF and the negative control being cells treated only with EGF (Fig. 2a). The coefficient of variation (CV), signal/background (S/B), and Z’-factor
were respectively defined as the ratio of the standard deviation (SD) and average of the controls, the ratio between the averages of the positive (signal) and negative (background) controls,
and the following equation: $${Z}^{{\prime} }=1-\left(3\times {{SD}}_{{positive}}+3\times {{SD}}_{{negative}}\right)/\left({{Avg}}_{{positive}}-{{Avg}}_{{negative}}\right)$$ (1) where
_SD__POSITIV__e_, _SD__negative_, _Avg__POSITIV__e_, and _Avg__negative_ represent the SD and average for the positive and negative controls, respectively. The obtained indices for our
system were 6% for CV, 6.2 for S/B, and 0.69 for Z’-factor. All these values were shown to satisfy the thresholds of ≤ 10% for CV, ≥2 for SB, and ≥ 0.50 for Z’-factor for drug screening
(Supplementary Table 1). Next, we carried out the screening using a library of 1134 FDA-approved compounds whose target proteins are responsible for broad functions such as receptors,
channels, kinases, enzymes, transporters, and so on (Fig. 2b, Supplementary Table 2, and Supplementary Data 1). This library includes 7 marketed drugs that act as TKI for EGFR (afatinib,
erlotinib, OSI-420 (the active metabolite of erlotinib), gefitinib, lapatinib, lapatinib ditosylate (another form of lapatinib), and vandetanib). To explore compounds significantly affecting
EGFR mobility, the screening procedure was designed as follows: 1) cultured cells in each well of a 96-well plate were pretreated with 100 μL of 10 μM compound solution at 37 °C for 1 h, 2)
single-molecule imaging was automatically done on 20 different cells, 3) 100 μL of 120 nM EGF solution was added by the automatic dispenser so that the final concentration of EGF was 60 nM,
and 4) another 20 cells were imaged 2 min after the EGF addition. During the screening, the Z’-factor was calculated for every assay plate from positive- and negative control wells (20
cells in each well) to evaluate the screening quality and images acquired from the plates passing the quality inspection were used in the following analysis (Supplementary Fig. 3a). To
achieve a highly accurate screening, MSDΔt was determined for every plate with the time duration (Δt) that provides the highest Z’-factor. For each compound, when the MSDΔt ratio was beyond
the average +3 SD of the MSDΔt ratio for the negative control, the compound was regarded as causing a significant MSD change of EGFR and identified as a hit compound. Figure 2c shows the
selected 53 compounds with MSDΔt ratios for compounds normalized to the negative control ratio. One of the marketed EGFR-targeted drugs, genistein, which is a flavonoid and known to inhibit
tyrosine kinases including EGFR, was excluded due to its IC50 (50% inhibitory concentration) being above 10 μM, which was beyond our experimental condition. For a more detailed assessment of
the selected 53 compounds, we measured the dose-dependent effects of the compounds on EGFR mobility with and without EGF (Supplementary Fig. 4 and Supplementary Data 2). Compounds with
significantly different MSD values at the minimum and maximum concentrations were selected. When half of the difference between the MSD (averaged over ~30 cells) at the two concentrations
exceeded the sum of their SD, the compound was regarded as a hit compound. As a result, 18 compounds were obtained, but one of them, auranofin, was excluded from further analysis due to its
cytotoxicity (cell death) during the treatment. Although these compounds showed similarly high MSD ratios, we found two types of effects on the MSD: one changed the receptor mobility
primarily after the EGF addition (Fig. 2d), and the other before the addition (Fig. 2e). Among the 17 compounds, 10 compounds corresponded to the former case and suppressed the EGF-dependent
decrease in a dose-dependent manner (Fig. 2d). These 10 were all kinase inhibitors: afatinib, erlotinib, OSI-420, gefitinib, lapatinib, and lapatinib ditosylate are known as EGFR-TKIs8,22;
ponatinib and vandetanib are pan-TKIs23; and dasatinib and ibrutinib have been reported as Bruton’s tyrosine kinase inhibitors24 and can interfere with EGFR phosphorylation. All these
EGFR-TKIs in the library were hit compounds (Supplementary Data 1), proving that our method was valid for exploring drugs targeting tyrosine phosphorylation of the receptor. The other 7 hit
compounds did not affect the EGF-induced decrease in mobility of EGFR but did affect the mobility without EGF stimulation (Fig. 2e). The reported drug actions and approved applicable
symptoms are not apparently concerned with EGFR tyrosine kinases: broxyquinoline is an anti-infective-agent; daunorubicin is an anthracycline antibiotic used for chemotherapeutic agents
against cancer by inhibiting DNA topoisomerase II25; eltrombopag is used to treat severe aplastic anemia and activates thrombopoietin receptor (TPOR) to facilitate platelet production26;
glafenine is a non-steroidal anti-inflammatory drug (NSAID) that inhibits cyclooxygenase-2/prostaglandin E2 (COX-2/PGE2) signaling27; nilotinib and sorafenib are not EGFR-TKI but TKI that
largely inhibit EGFR-independent pathways and used respectively to treat primary kidney cancer28 and chronic myeloid leukemia29; and, finally, curcumin interferes with tyrosine kinases
including EGFR and is approved as a food additive. Because these compounds induced EGFR mobility changes without EGF, they are suggested to interact directly or indirectly with EGFR by some
actions other than the inhibition of tyrosine kinase. The effects of these compounds on EGFR are described in more detail below. EGFR CLUSTERING-BASED SCREENING Single-molecule tracking can
obtain the brightness of each EGFR-mEGFP fluorescent spot, which reflects the number of fluorescent GFP involved. Uniform illumination was required to quantify the brightness. AiSIS achieved
uniform TIR illumination over 3000 μm2. Because CHO-K1 cells express no endogenous EGFR, their brightness typically reflects the oligomer/clustering size. The EGF concentration correlated
with the brightness (Fig. 3a and b), indicating that monomers/dimers were relatively reduced with increasing concentration, and larger size clusters were gained. The formation of clusters
with integrating monomers/dimers has been suggested to correlate with the activation of downstream signaling through interactions with adapter molecules15. Thus, the increase in brightness
could be used as a screening index to assess signal transduction. For the obtained single-molecule tracking data, we compared the probability density distributions of the spot brightness
before and after compound treatment without EGF. The Kullback–Leibler divergence (KLD) was used to quantify the similarity of the histogram profiles. In this case, Z’-factors were calculated
using KLD before and after the EGF addition (without any compound) as respective negative and positive controls. The observed Z’-factors did not satisfy the threshold of 0.5 due to the
broad SD (Supplementary Fig. 3b). Nevertheless, the highest KLD value among the compounds (Fig. 3c) was obviously different from the KLD calculated for the negative control. This compound,
verteporfin, diminished the number of brighter spots in a dose-dependent manner (Fig. 3d–f), suggesting that larger clusters of EGFR decreased on the plasma membrane. Verteporfin reduced
EGFR diffusion in a dose-dependent manner without EGF, although it was not selected from the mobility-based screen due to its modest effects (Fig. 3g). Verteporfin is a photosensitizing drug
for age-related macular degeneration30. As described in more detail below, the effect of this compound on EGFR was due to EGFR internalization and not to any photophysical effect.
CHARACTERIZATION OF HIT COMPOUNDS ON EGFR DYNAMICS, SIGNAL TRANSDUCTION, AND CELL VIABILITY To determine whether these non-EGFR-TKI compounds affect EGFR-dependent signaling in CHO-K1 cells
expressing EGFR, which were used in the screening, we examined the activation and expression levels of both EGFR and the downstream signaling protein ERK (Fig. 4a–c). Treatment with any of
five non-EGFR-TKI compounds (broxyquinoline, daunorubicin, eltrombopag, sorafenib, and verteporfin) caused no significant inhibition of EGFR phosphorylation upon EGF stimulation in
comparison with the control (compound-untreated) condition (Fig. 4a and b, left), a finding consistent with these compounds not being EGFR-TKIs. In the subsequent signaling, the
phosphorylation of ERK fell upon treatment with any of the five compounds, especially eltrombopag, sorafenib, and verteporfin, although the expression levels of ERK were not affected,
indicating an inhibition of EGFR-dependent signaling (Fig. 4a and b, right). The total amount of EGFR was decreased by treatment with the five compounds (Fig. 4c) regardless of EGF
stimulation, indicating the destabilization of EGFR. Furthermore, EGFR was internalized in the cells treated with the five compounds without EGF, as shown in timelapse TIRF images (Fig. 4d
and Supplementary movie 3) and by quantification (Fig. 4e). In the case of verteporfin, the photoreactive damage of GFP induced by repetitive laser irradiation was observed because of the
photosensitizing effect of verteporfin (Supplementary Fig. 5a). Further observations of EGFR-mEGFP at the condition minimizing the photophysical effects suggested EGFR internalization (Fig.
4d, e, and Supplementary Fig. 5b, c). The examination by immunofluorescence microscopy of verteporfin supported EGFR internalization, as described later. In contrast with these five
non-EGFR-TKI compounds, when treated with the other two compounds, glafenine and nilotinib, the cells exhibited no reduction of EGF-induced EGFR phosphorylation or downstream ERK (Fig. 4b)
and showed no obvious internalization or degradation of EGFR (Fig. 4c and e). Thus, the five non-EGFR-TKI compounds (broxyquinoline, daunorubicin, eltrombopag, sorafenib, and verteporfin)
caused the downregulation of EGFR-related signal transduction with internalization, even though EGFR phosphorylation was unchanged. That is, compounds that can alter EGFR signaling outside
tyrosine phosphorylation were successfully selected by the single-molecule tracking-based screening method. We next evaluated the effects of the hit compounds on EGFR-dependent cellular
viability by utilizing various cell types that express EGFR because they are expected to depend on EGFR signaling for their survival: A431 and HeLa cells, which express endogenous EGFR, and
EGFR-transfected Ba/F3 cells. We also utilized Ba/F3 and CHO-K1 cells, which do not express EGFR, as controls. Viability assays were carried out under 10 μM of compound for 72 h. The left
panel in Fig. 5a shows that all hit EGFR-TKIs (i.e., afatinib, erlotinib, gefitinib, lapatinib, ponatinib, vandetanib, dasatinib and ibrutinib) reduced the viability of A431 cells and
EGFR-transfected Ba/F3 cells, consistent with a dependency on EGFR activity for their survival31,32. HeLa cells, which expressed EGFR 10-fold less than A431 cells33, also exhibited less
viability with EGFR-TKI treatment, although they were somehow resistant to erlotinib and gefitinib, consistent with previous reports34,35,36. Similar to the effects of most EGFR-TKIs, the
five non-EGFR-TKI compounds (broxyquinoline, daunorubicin, eltrombopag, sorafenib, and verteporfin) suppressed the viability of the three EGFR-expressing cell types, while the other two
non-EGFR-TKIs (glafenine and nilotinib) caused higher viability. In the right panel in Fig. 5a, the viability of CHO-K1 and Ba/F3 cells was not significantly suppressed by EGFR-TKIs or the
five non-EGFR-TKI compounds. These observations indicate that the five non-EGFR-TKI compounds suppress EGFR-dependent cell survival. The viability of CHO-K1 cells lacking EGFR expression was
sensitive to ponatinib and dasatinib, possibly due to these compounds acting as TKIs for other tyrosine kinases as well as EGFR. We further evaluated the effects of the five non-EGFR-TKI
compounds on EGFR signaling in the various cell lines. The EGF-induced phosphorylation of EGFR was observed with the non-EGFR-TKI compounds in A431, HeLa, and EGFR-transfected Ba/F3 cells
(Fig. 5b and Supplementary Fig. 6), although at varying degrees, especially for verteporfin. The expression level of EGFR tended to be reduced slightly by the five compounds in A431 and
EGFR-transfected Ba/F3 cells but rarely in HeLa cells (Fig. 5c). Verteporfin, in particular, caused EGFR-destabilization in EGFR-transfected Ba/F3 cells, which can explain the reduced
EGF-induced EGFR phosphorylation (Fig. 5b). The stability and internalization of EGFR were further examined in the EGFR-expressing cell lines by immunofluorescence microscopy, in which the
photophysical effect of verteporfin need not be considered. Treatment with any of the five non-EGFR-TKI compounds caused a decrease in EGFR localization on the membrane in A431, HeLa, and
EGFR-transfected Ba/F3 cells compared to untreated cell lines (Fig. 5d). In particular, eltrombopag, sorafenib, and verteporfin tended to decrease the total expression level of EGFR,
suggesting the internalization and degradation of EGFR. Figure 5e shows the ratio of EGFR localization on the plasma membrane and in the cytoplasm of the EGFR-positive cell lines treated
with the non-EGFR-TKI compounds, revealing the five compounds enhanced the internalization of EGFR. In contrast, the other two compounds (glafenine and nilotinib) did not alter
EGFR-dependent cell viability (Fig. 5a), EGF-induced EGFR phosphorylation (Fig. 5b), the expression level of EGFR (Fig. 5c), or the membrane localization of EGFR (Fig. 5d and e), indicating
no effects on EGFR-dependent signaling or cell survival, although these compounds affected EGFR mobility (Fig. 2e). Thus, the hit compounds, which were selected using EGFR-expressing CHO-K1
cells, were confirmed to robustly exert inhibitory effects on the EGFR-dependent cell viability by inducing the internalization and degradation of EGFR in various cell types including A431,
EGFR-transfected Ba/F3, and Hela cells, albeit to varying degrees. Overall, the hit EGFR-TKIs that inhibited the phosphorylation-dependent mobility shift of EGFR reduced the EGFR-dependent
viability. The five non-EGFR-TKI compounds (broxyquinoline, daunorubicin, eltrombopag, sorafenib, and verteporfin) that decreased EGFR diffusion or clusters without EGF, reduced
EGFR-dependent cell viability by downregulating EGFR. In contrast, the other two non-EGFR-TKI compounds (glafenine and nilotinib) that affected EGFR mobility without EGF stimulation did not
affect the EGFR-dependent signaling needed for cell survival. DISCUSSION Here, we demonstrated single-molecule tracking-based drug screening to explore compounds effective against EGFR. In
addition to the methods used in previous single-molecule screening for nuclear receptors37, AiSIS enabled highly precise auto-focusing on the plasma membrane receptor and uniform TIR
illumination for the quantification of fluorescence brightness. Our method observed changes in EGFR mobility and clustering instead of focusing on a specific process of the signaling,
allowing it to detect changes in multiple processes dependent on the ligand. Because changes in the mobility and cluster distribution of the target molecule can hardly be measured by
biochemical methods but can be observed by single-molecule imaging as signaling processes, our single-molecule tracking-based screening has the potential to detect compounds that cannot be
detected by conventional methods. Screening by assessing several treatment conditions of ligand/compound led to a wide selection of compounds affecting EGFR functions that are related or
unrelated to EGF stimulation (Fig. 2). Among the hit compounds, those suppressing the EGF-induced mobility shift of EGFR were identified as EGFR-TKIs, proving the reliability of the method
for drug screening. One of the possible reasons why EGFR-targeted TKIs can be detected by mobility-based screening is described in Fig. 6a. Accumulating evidence shows that EGFR undergoes
structural changes during its phosphorylation upon EGF stimulation and then translocates to a confined membrane region via interactions with membrane components (lipid, proteins, etc.),
leading to slower receptor mobility15. Changes in the MSD of EGFR were proven to couple tightly with the phosphorylation in a dose-dependent manner (Fig. 1f). The competitive docking of TKI
to the ATP-binding pocket of EGFR prevents the receptor from subsequent structural changes during phosphorylation, which thus suppresses the decrease in mobility and subsequent signaling
processes. In addition to the detection of EGFR-TKIs, our screening detected non-EGFR-TKI compounds (i.e., broxyquinoline, daunorubicin, eltrombopag, sorafenib, verteporfin, glafenine, and
nilotinib), none of which are known to act on EGFR directly. Figure 6b illustrates a scheme for the broxyquinoline, daunorubicin, eltrombopag, sorafenib, and verteporfin effects; these
compounds commonly reduced mobility, induced internalization, decreased the expression levels of EGFR, and affected EGFR-dependent signaling and viability. We assume that these compounds
force EGFR to a specific membrane subdomain where receptor mobility is lower. EGFR in the membrane subdomains may internalize via caveolin-mediated endocytosis38,39. The internalization
exerts a harmful effect on cell viability by suppressing EGFR signaling for proliferation and survival40. For example, oxidative processes in cells are induced by various compounds (e.g.,
broxyquinoline) and have been suggested to affect EGFR signaling41. Previous studies have shown that broxyquinoline stabilizes hypoxia-inducible factor (HIF)−142 to accumulate caveolin-1 in
the lipid raft to constitute caveolae43, which directly interacts with EGFR, suggesting the confined mobility and internalization of the protein complex including EGFR. In fact, we confirmed
that broxyquinoline increased HIF-1 and the internalization of EGFR with caveolin in EGFR-transfected CHO-K1 cells (Supplementary Fig. 7). Furthermore, some compounds may induce complex
formation including EGFR, thereby promoting the internalization and degradation of EGFR38,44,45. In addition, some compounds are reported to interact with the plasma membrane to possibly
alter EGFR mobility46,47. Figure 6c illustrates the case of glafenine and nilotinib. No significant change was observed in the EGFR-dependent viability of cells treated by these two
compounds (Fig. 5). Additionally, the phosphorylation of downstream proteins was not suppressed, and no internalization/reduction of EGFR expression occurred (Figs. 4 and 5), indicating no
downregulation of EGFR signaling by these two compounds. As described above, the hit compounds obtained in the present study were categorized into three types based on the observed EGFR
behavior and cell responses, with each type suggested to depend on a different mechanism of action. The scheme in Fig. 6a, which illustrates that EGFR-TKIs can be obtained by the
mobility-based screening, is useful for searching for effective TKIs if EGFR mutants resistant to existing TKIs appear to change their motility upon EGF-stimulated activation48. The scheme
in Fig. 6b, which was suggested from the five non-EGFR-TKI compounds, is effective for removing pathogenic cells that excessively express EGFR, a common phenotype of various cancers, by
inducing cell death following EGFR internalization. Therefore, drug repositioning for EGFR drugs may be possible using compounds that exhibit this scheme. In Fig. 6c, compounds that do not
significantly affect cell viability but exert an effect on receptor mobility might be drug seeds with little cytotoxicity. Besides EGFR, various types of receptors on the plasma membrane
have been reported to change their mobility according to their activation and the cluster formation to propagate downstream signaling15,37,49, suggesting that single-molecule tracking-based
drug screening is applicable to a wide variety of membrane receptors. METHODS CELL PREPARATION CHO-K1, A431, and HeLa cells (RIKEN BRC through the National Bio-Resource Project of the MEXT,
Japan) were grown at 37 °C in Ham’s F-12 medium (CHO-K1) or Dulbecco’s modified Eagle’s medium (A431, HeLa; DMEM, FUJIFILM Wako Pure Chemical, Japan); both media were supplemented with 10%
fetal bovine serum (FBS). Ba/F3 cells (RIKEN BRC) and Ba/F3 cells expressing EGFR50,51 (a kind gift from Dr. Ryo Iwamoto and Dr. Eisuke Mekata, Osaka University) were cultured at 37 °C in
RPMI medium supplemented with 10% FBS and 4 ng/mL mouse IL3 (091-03971, Fuji-Wako, Japan). In the case of Ba/F3 cells expressing EGFR, 20 ng/mL EGF (315-09, PeproTech, USA) was added to the
medium. The culture medium was replaced with DMEM minus phenol red or FBS for starvation two hours before single-molecule imaging when the medium was changed to DMEM containing 5 mM PIPES.
The cells were cultured until 90% confluence and observed in 60 wells of a 96-well plate (GP96000, Matsunami Glass, Japan); the peripheral wells were excluded to avoid interference between
the microscope stage and the objective lens. A stable CHO-K1 cell line expressing EGFR-mEGFP with an appropriate molecular density (1–3 μm−2) on the basal membrane was established for
single-molecule tracking. A DNA construct linking the cDNA of human EGFR (Supplementary Data 3, provided by Akihiko Yoshimura, Keio University) to the pEGFP-C1 vector (Clontech, Japan) was
transfected to parental CHO-K1 cells with 80% confluency in a 6 cm dish using FuGENE HD (Promega, USA) with 2 µg of the construct mixed with 500 µL of OptiMEM (31985-062, Gibco, USA). A
fraction of cells expressing EGFR-mEGFP was collected using a SH800S cell sorter (Sony, Japan) 2–3 days after the transfection. By sorting with a 488 nm laser, cells exhibiting a higher
fluorescence intensity than the autofluorescence measured beforehand were selected and isolated. Cell cloning was carried out using the limiting dilution method, and the expression level of
EGFR-mEGFP was confirmed for each clone using TIRFM by determining if individual fluorescent spots were distinguishable. When the expression level of EGFR-mEGFP was considerably increased
during the passage, the cell sorting and cloning process were executed again to supply the appropriate cells to single-molecule imaging. AI-aided cell-searching was applied to the imaging to
acquire images of cells with a suitable molecular density of 0.76 ± 0.55 μm−2. AUTOMATED IN-CELL SINGLE-MOLECULE IMAGING SYSTEM (AISIS) AiSIS consists of an automated microscope for
single-molecule imaging and robotics for liquid dispensing. Total internal reflection (TIR) illumination was configurable with a high-magnification objective, PlanApo 60X NA 1.49 (Nikon,
Japan), to acquire single-molecule images at the basal cell surface under an inverted microscope (Ti2-E, Nikon, Japan). Auto Imaging System (ZIDO Corp., Japan), including an autofocus device
and AI-aided cell searching applications, was equipped to automatically obtain in-focus images of the cells with a suitable density of fluorescent spots for single-molecule analysis. Lasers
at wavelengths of 488 nm (OBIS 488LS, Coherent, USA) were used to excite GFP fused with EGFR. The dichroic mirror/emission filter set was DM495/BA500-545 (Nikon, Japan). The images were
acquired at a frame rate of 33 ms for 25 frames using a sCMOS camera (ORCA-Flash4.0 V2, Hamamatsu, Japan). A liquid-handling robot (Cavro Omni Robot, Tecan, USA) automatically sucked 100 μL
of two-fold the final concentration of EGF solution from the well in the storage plate and dispensed the solution into the target well of the cell culture plate with 100 μL of observation
medium. AUTOFOCUS DEVICE The device consists of a light source, magnifier optics, a slit located at the plane optically conjugated to the basal cell surface, a CCD camera for acquiring the
slit image, and a control unit that feedback-controls the position of the objective lens (Fig. 1b). An 830 nm laser is used as the light source, and its incident angle is switched between
two values using a Galvano mirror with a frequency of 10 Hz. The slit image on the CCD is moved according to the position of the objective lens. When the objective is located at an under- or
over-focus position, the image is seen at the opposite side across the center, where it is formed by the objective at the in-focus position (Supplementary Fig. 1a). The center can be
identified quickly as the middle point between two different slit images obtained by switching the incident angle of the laser 180 degrees. The side of the image corresponds to the
z-direction of the shift of the objective from the in-focus position. The calculated deviation from the center is transmitted to a control unit to feedback-control the objective position.
The gap difference between the in-focus positions determined by the device and human eyes can be adjusted by setting an offset value. Supplementary Fig. 1b shows the flow of the autofocusing
process. This method, using two images with light from exactly opposite directions, is robust for variations in the sample condition (e.g., refractive index) that affect the optical path at
the sample-coverslip interface and cause inaccurate focusing by conventional methods. The device can also be applied to normal microscopy, especially with high magnification. In
Supplementary Fig. 1c, another method for switching the laser direction is introduced. A circular transparent plate with a thin circular coating (e.g., aluminum) forms thin
light-nontransparent regions inside and outside each half of the circle. The laser illumination goes through the circles; therefore, when the plate rotates, the optical path is switched
according to the rotational frequency, enabling illumination from opposite directions. SINGLE-MOLECULE TRACKING Single-molecule tracking was carried out on the acquired images to obtain the
positions and brightness of fluorescent EGFR-mEGFP using commercial software (Auto Analysis Software, AAS, ZIDO, Japan). The first process of the tracking was to determine bright spots in
the image by selecting square regions with 11 × 11 pixels beyond a threshold for cross-correlation between pixel values and the two-dimensional Gaussian distribution. Then, the spot region
was fitted with the following function: $$I\left(x,y ;\, {I}_{0},{x}_{g},{y}_{g},{\sigma }_{A},a,b,{I}_{{back}}\right)= {I}_{0}\,
{exp}\left[-\frac{{(x-{x}_{g})}^{2}+{(y-{y}_{g})}^{2}}{2{\sigma }_{A}^{2}}\right]\\ +a\left(x-{x}_{g}\right)+b\left(y-{y}_{g}\right)+{I}_{{back}},$$ (2) where _x_ and _y_ denote the pixel
positions. The distribution of the pixel values was expressed as a Gaussian function with a peak intensity of _I__0_ at the centroid, (_x__g_, _y__g_) and a variance of _σ__A_2, plus a
background with an inclination described by _a_ and _b_ above the offset intensity _I__back_. A single-molecule trajectory was generated by connecting the spots as follows: all possible
connections between two spots at times _t_ and _t_ − 1 with a center-to-center distance below 6 pixels were listed, and the shortest connection was selected. Trajectories outside the cell
region determined by the AI-aided cell search algorithm were removed. MOLECULAR MOBILITY ANALYSIS The mobility of lateral diffusion was analyzed with the MSD calculated from the positions of
the fluorescent spots with the following equation: $${MSD}\left(n \Delta t\right)={\left[{\left\{{x}_{i}\left(n\Delta t+m\Delta t\right)-{x}_{i}(m\Delta
t)\right\}}^{2}+{\left\{{y}_{i}\left(n\Delta t+m\Delta t\right)-{y}_{i}(m\Delta t)\right\}}^{2}\right]}_{i,m}$$ (3) Here, _x__i_ and _y__i_ represent the single-molecule position in the i-th
track, _n_ and _m_ denote the frame number, _Δt_ is the time interval between frames (33 ms), and []_i,m_ denotes the average over _i_ tracks and _m_ frames. For Fig. 1f and g, MSD at Δt =
500 ms was used. The dose-response curve (Fig. 1f) was plotted as the MSD ratio and fitted using the following equation to calculate the EC50: $${MSD} \,=\, {{MSD}}_{\max
}-\frac{{{MSD}}_{\max }-{{MSD}}_{\min }}{1+{\left(\frac{{{EC}}_{50}}{\left[L\right]}\right)}^{h}},$$ (4) where _MSD__max_ and _MSD__min_ are the MSD ratios of the upper and lower boundaries,
respectively, [_L_] is the ligand (EGF) concentration, and _h_ is the Hill coefficient. To display the dose-response curve in comparison with the phosphorylation level, the MSD ratio was
converted as follows: $${{MSD}}_{{converted}}=\frac{{{MSD}}_{\max }-{MSD}}{{{MSD}}_{\max }-{{MSD}}_{\min }}$$ (5) COMPOUNDS The library of FDA-approved compounds (Selleck Chemicals, USA) was
provided in 96-well plates from the Center for Supporting Drug Discovery and Life Science Research, Osaka University, in 2021. 3 μL of 1 mM compound in DMSO was dispensed in each well and
diluted to 10 μM with DMEM before screening. The medium in 60 wells of the 96-well plate with cultured cells was replaced to the compound solution and incubated at 37 °C for 1 h. Both sides
of the compound-treated wells (total 2 × 6 wells) were used for the positive and negative controls (with and without 10 μM gefitinib), respectively. SINGLE-MOLECULE SCREENING For the
compound screening, single-molecule imaging was executed on 20 cells before and 20 cells after EGF stimulation. 100 μL of 120 nM EGF was added to every well (final concentration, 60 nM). The
MSD ratio was used to confirm the quality of the screening and select the hit compounds. For each plate, we calculated the Z’-factor using the MSD ratios of the wells with the positive (10
µM gefitinib in DMSO) and negative (DMSO only) controls. The MSD at Δt that provided the best Z’-factor in each well plate was used for Fig. 2c and at 167 ms for Fig. 2d and e. The average
and SD of the MSD ratio of positive and negative controls were used to calculate the Z’-factor. $${Z}^{{\prime} }=1-\frac{\left(3\times {{SD}}_{{positive}}\right)+\left(3\times
{{SD}}_{{negative}}\right)}{{{Avg}}_{{positive}}-{{Avg}}_{{negative}}}$$ (6) Here, _Avg_ and _SD_ respectively represent the averages and SD of the MSD for the positive and negative
controls. In Fig. 2c, the MSD ratio for each compound was normalized to that of the negative control, and any compound with a ratio greater than the sum of the average and three-fold SD of
the negative control was defined as a hit compound. For the compound-dose dependent assay shown in Fig. 2d and e, cells were treated with 100 µL of 10 µM, 1 µM, 0.1 µM, 0.01 µM, and 0.001 µM
compound solution for 1 h before single-molecule imaging. Regardless of the EGF addition, when the MSD value was larger than half the difference of the MSD at the minimum and maximum
compound concentration plus their SD, the compound was recognized as effective for EGFR. CELL VIABILITY ASSAY A431, HeLa, Ba/F3 with and without EGFR expression, and CHO-K1 cells in 96-well
plates (1860-096, Iwaki, Japan) were incubated in 10 μM compound solution at 37 °C for 72 h. Compound solutions for the A431, HeLa, and CHO-K1 cells were prepared by diluting stock solutions
of compounds in HBSS. The solutions additionally contained EGF but no IL-3 for Ba/F3 cells expressing EGFR and only IL-3 for Ba/F3 cells not expressing EGFR. Then, the medium was replaced
with 10 μL of Cell Counting Kit −8 (Dojindo, Japan) diluted in 100 μL HBSS. After 2 h of incubation, absorption of the medium was measured for each well with a wavelength of 450 nm using a
plate reader (Infinite F50 Plus, Tecan, USA). WESTERN BLOTTING After the compound and/or EGF treatment, the cells were lyzed in SDS sample buffer. The lysate was electrophoresed on a 10%
SDS-polyacrylamide precast gel (192-14961, SuperSep Ace, 10%, 17well, FUJIFILM Wako Pure Chemical, Japan) Then, the proteins were transferred to a 0.45 μm pore PVDF membrane (034-25663,
ClearTrans PVDF Membrane, Hydrophobic, 0.45 µm, FUJIFILM Wako Pure Chemical, Japan) and reacted with antibodies against the following targets: EGFR (#4267, Cell signaling technology (CST),
USA), phospho-EGFR (Tyr1068, #4267, CST, USA), AKT (#9272, CST, USA), phospho-AKT (Ser473, #4060, CST, USA), ERK1/2 (#9107, CST, USA), and phospho-ERK1/2 (Thr202/Try204, #9106, CST, USA).
For chemiluminescent antibody detection, HRP-linked anti-rabbit IgG (#7074, CST, USA) and HRP-linked anti-mouse IgG (#7076, CST, USA) were used with ECL prime reagent (Cytiva, USA). The
loading control containing proteins ranging from 37 to 250 kDa was obtained from the SDS gel stained with CBB. Quantification of the band densities was carried out using ImageJ software
(NIH). Two squares were set such that one square was placed in the band and another square was far away from the band, and the difference in the average intensities of these regions was
defined as the band intensity. The intensity of the anti-phosphorylated antibody was normalized to that of the anti-protein antibody. The phosphorylation level was calculated by dividing the
normalized intensity of the phosphorylated protein for each condition by that for 30 nM EGF without compound. The EGF dose-response curve was fitted with the Hill equation as follows:
$${phosphorylation}=\min -\frac{\max -\min }{1+{\left(\frac{{{EC}}_{50}}{\left[L\right]}\right)}^{h}}.$$ (7) Here, _h_ indicates the Hill coefficient, _max,_ and _min_ denote upper and lower
bounds, respectively, and [_L_] is the concentration of the ligand, EGF. The protein expression amount was calculated by normalizing the intensity of the protein for each condition by that
of the loading control. INTERNALIZATION ASSAY For quantification of the internalization (Fig. 4d and e), CHO-K1 cells expressing EGFR-mEGFP in a 96-well plate were treated in compound
solution for 1 h. The compound solutions were the same as those used in the experiment above. After the compound treatment, timelapse single-molecule images (Fig. 4d) were acquired by AiSIS
at the same positions in a well every 10 min. For verteporfin, which showed rapid photobleaching of EGFR-mEGFP as a photophysical effect induced by repetitive laser irradiation, the images
were obtained at the same positions at only two-time points (0 min and 20, 40, 60, or 80 min) to minimize the effect. In the obtained images, the average brightness was measured for circled
regions of interest (ROI) within a cell and outside the cell (background). The difference in the brightness between these regions was calculated for each time point and normalized to the
value at time 0 (Fig. 4e). To consider the effect of fluorescence bleaching caused by repetitive irradiation by the laser, timelapse images of the cells without compound treatment were
acquired and analyzed by the same method. IMMUNOFLUORESCENCE MICROSCOPY The compound treatment of A431 and HeLa cells cultured in 24-well plates was carried out in 10 μM solutions for 1 hour
at 37°C after the cells were incubated in DMEM without FBS for 6 hours. Subsequently, the compounds were washed out with HBSS, and the cells were fixed with 4% PFA for 30 min at 4 °C and
solubilized with 0.5% Triton X100 in HBSS at room temperature, followed by washing with HBSS three times and left at 4 °C overnight. After the blocking process with HBSS containing 2% BSA
for 15 min, 1:300-diluted anti-EGFR antibody (#4267, CST, USA) or 1:10,000-diluted anti-caveolin antibody (#3267 T, CST, USA) was added as the primary antibody for 1 hour at room
temperature. The cells were washed with HBSS three times and labeled with 1:1000 anti-IgG antibody conjugated with Alexa 488 or Alexa 647 (#A-11034 or #A-21244, respectively, Invitrogen,
USA) for 1 h at room temperature. The nucleus was fluorescently labeled with 0.05% NucSpot Live 650 (Biotium, USA) for 30 min at room temperature. Observation of the samples was done by
confocal microscopy (Nikon A1) with a 20X objective lens (Nikon PalnApo) operated with NIS elements software (Nikon). The preparation process for EGFR-transfected Ba/F3 cells was almost the
same as that for other cells, except that floating cells in Eppendorf tubes were centrifuged with 800 × _g_ for 2 min to replace the supernatant after every step. For the observation, the
cells were suspended in HBSS in a 96-well plate. The obtained images were analyzed by quantifying the fluorescence intensity of the plasma membrane and cytoplasm to calculate the ratio of
EGFR in these regions. The regions of the plasma membrane and cytoplasm were identified as the area within 5 pixels inside the cell boundary, which was determined with Cellpose 3.0, and the
other pixels within the cell, respectively. The average fluorescence intensities of the plasma membrane (_F_mem), the cytoplasm (_F_cyt), and background area (_F_bck) where no cells existed
were obtained to calculate the ratio, _I_mem_/I_cyt = (_F_mem − _F_bck)/(_F_cyt − _F_bck), as the extent of internalization. In addition, the decrease in total EGFR in a cell due to
degradation was taken into account as the intensity ratio of the whole cell with to without compound treatment _α_ = (_F_drug − _F_bck)/(_F_ctrl − _F_bck). The product of this ratio (α_・
I_mem_/I_cyt) includes both the internalization and degradation effects of a compound, which correspond to the fluorescence change in the obtained image. The calculations were performed
using a custom-made program written in Python 3.8.18. REPORTING SUMMARY Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
DATA AVAILABILITY Source data are provided with this paper. All other data supporting the findings of this study are available in the Supplementary Information file and from the
corresponding author upon request. CODE AVAILABILITY All Python and ImageJ scripts supporting the findings of this paper are available upon reasonable request. The code used in this paper is
deposited in the GitHub repository at https://github.com/d-watana/Single-Molecule-Imaging-Data-Analysis. REFERENCES * Santos, R. et al. A comprehensive map of molecular drug targets. _Nat.
Rev. Drug Discov._ 16, 19–34 (2016). Article PubMed Google Scholar * Hopkins, A. L. & Groom, C. R. The druggable genome. _Nat. Rev. Drug Discov._ 1, 727–730 (2002). Article CAS
PubMed Google Scholar * Vasaikar, S., Bhatia, P., Bhatia, P. G. & Yaiw, K. C. Complementary approaches to existing target based drug discovery for identifying novel drug targets.
_Biomedicines_ 4, 27 (2016). Article PubMed Google Scholar * Kovacs, T., Zakany, F. & Nagy, P. It takes more than two to tango: complex, hierarchal, and membrane-modulated
interactions in the regulation of receptor tyrosine kinases. _Cancers_ 14, 944 (2022). Article CAS PubMed Google Scholar * Normanno, N., Maiello, M. R. & Luca, A. D. E. Epidermal
growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs): simple drugs with a complex mechanism of action? _J. Cell. Physiol._ 19, 13–19 (2002). Google Scholar * Roengvoraphoj, M.,
Tsongalis, G. J., Dragnev, K. H. & Rigas, J. R. Epidermal growth factor receptor tyrosine kinase inhibitors as initial therapy for non-small cell lung cancer: Focus on epidermal growth
factor receptor mutation testing and mutation-positive patients. _Cancer Treat. Rev._ 39, 839–850 (2013). Article CAS PubMed Google Scholar * Chiba, M. et al. Efficacy of irreversible
EGFR-TKIs for the uncommon secondary resistant EGFR mutations L747S, D761Y, and T854A. _BMC Cancer_ 17, 281 (2017). Article PubMed PubMed Central Google Scholar * Du, X. et al. Acquired
resistance to third-generation EGFR-TKIs and emerging next-generation EGFR inhibitors Acquired resistance to third-generation EGFR-TKIs and emerging next-generation EGFR inhibitors.
_Innovation_ 2, 100103 (2021). CAS PubMed PubMed Central Google Scholar * Gao, J., Jian, J., Jiang, Z. & Van Schepdael, A. Screening assays for tyrosine kinase inhibitors: a review.
_J. Pharm. Biomed. Anal._ 223, 115166 (2023). Article CAS PubMed Google Scholar * Wee, P. & Wang, Z. Epidermal growth factor receptor cell proliferation signaling pathways. _Cancers_
9, 52 (2017). Article PubMed PubMed Central Google Scholar * Peters, C. et al. Characterization of a new molecule capable of inhibiting several steps of the amyloid cascade in
Alzheimer’s disease. _Neurobiol. Dis._ 141, 104938 (2020). Article CAS PubMed Google Scholar * Sako, Y., Minoguchi, S. & Yanagida, T. Single-molecule imaging of EGFR signalling on
the surface of living cells. _Nat. Cell Biol._ 2, 168–172 (2000). Article CAS PubMed Google Scholar * Ueda, M., Sako, Y., Tanaka, T., Devreotes, P. & Yanagida, T. Single-molecule
analysis of chemotactic signaling in Dictyostelium cells. _Science_ 294, 864–867 (2001). Article ADS CAS PubMed Google Scholar * Iino, R., Koyama, I. & Kusumi, A. Single molecule
imaging of green fluorescent proteins in living cells: E-cadherin forms oligomers on the free cell surface. _Biophys. J._ 80, 2667–2677 (2001). Article CAS PubMed PubMed Central Google
Scholar * Hiroshima, M. et al. Transient acceleration of epidermal growth factor receptor dynamics produces higher-order signaling clusters. _J. Mol. Biol._ 430, 1386–1401 (2018). Article
CAS PubMed Google Scholar * Hiroshima, M., Saeki, Y., Okada-Hatakeyama, M. & Sako, Y. Dynamically varying interactions between heregulin and ErbB proteins detected by single-molecule
analysis in living cells. _Proc. Natl Acad. Sci. USA_ 109, 13984–13989 (2012). Article ADS CAS PubMed PubMed Central Google Scholar * Clarke, D. T. & Martin-Fernandez, M. L. A
brief history of single-particle tracking of the epidermal growth factor receptor. _Methods Protoc._ 2, 12 (2019). Article CAS PubMed PubMed Central Google Scholar * Byeon, H. K. Beyond
EGFR inhibition: multilateral combat strategies to stop the progression of head and neck cancer. _Exp. Mol. Med._ 51, 8 (2019). Article PubMed PubMed Central Google Scholar * Gamble, M.
C. et al. Mu-opioid receptor and receptor tyrosine kinase crosstalk: Implications in mechanisms of opioid tolerance, reduced analgesia to neuropathic pain, dependence, and reward. _Front.
Syst. Neurosci._ 16, 1059089 (2022). Article CAS PubMed PubMed Central Google Scholar * Yasui, M., Hiroshima, M., Kozuka, J., Sako, Y. & Ueda, M. Automated single-molecule imaging
in living cells. _Nat. Commun._ 9, 3061 (2018). Article ADS PubMed PubMed Central Google Scholar * Yasui, M., Hiroshima, M. & Ueda, M. United States Patent: 11567293. (2023). *
Abourehab, M. A. S., Alqahtani, A. M., Gouda, B. G. M. Y. & Gouda, A. M. Globally approved EGFR inhibitors: insights into their syntheses, target kinases, biological activities, receptor
interactions, and metabolism. _Molecules_ 26, 6677 (2021). Article CAS PubMed PubMed Central Google Scholar * Falco et al. Ponatinib (AP24534) is a novel potent inhibitor of oncogenic
RET mutants associated with thyroid cancer. _J. Clin. Endocrinol. Metab._ 98, 811–819 (2013). Article Google Scholar * Joseph, R. E. et al. Differential impact of BTK active site
inhibitors on the conformational state of full-length BTK. _eLife_ 9, e60470 (2020). Article CAS PubMed PubMed Central Google Scholar * Lehmann, M. et al. Activity of topoisomerase
inhibitors daunorubicin, idarubicin, and aclarubicin in the drosophila somatic mutation and recombination test. _Environ. Mol. Mutagen._ 257, 250–257 (2004). Article Google Scholar *
Grainger, J. D. Eltrombopag for the treatment of aplastic anemia: current perspectives. _Drug Des. Dev. Ther._ 10, 2833–2843 (2016). Article Google Scholar * Carlile, G. W. et al. The
NSAID glafenine rescues class 2 CFTR mutants via cyclooxygenase 2 inhibition of the arachidonic acid pathway. _Sci. Rep._ 12, 4595 (2022). Article ADS CAS PubMed PubMed Central Google
Scholar * Guevremont, C., Jeldres, C., Perrotte, P. & Karakiewicz, P. I. Sorafenib in the management of metastatic renal cell carcinoma. _Curr. Oncol._ 16, S27–S32 (2009). Article
PubMed PubMed Central Google Scholar * Kantarjian, H. M. et al. Nilotinib is effective in patients with chronic myeloid leukemia in chronic phase after imatinib resistance or intolerance:
24-month follow-up results. _Blood_ 117, 1141–1145 (2011). Article CAS PubMed PubMed Central Google Scholar * Borodoker, N. et al. Verteporfin infusion-associated pain. _Am. J.
Ophthalmol._ 133, 211–214 (2002). Article CAS PubMed Google Scholar * Janmaat, M. L., Kruyt, F. A. E., Rodriguez, J. A. & Giaccone, G. Response to epidermal growth factor receptor
inhibitors in non-small cell lung cancer cells: Limited antiproliferative effects and absence of apoptosis associated with persistent activity of extracellular signal-regulated kinase or Akt
kinase pathways. _Clin. Cancer Res._ 9, 2316–2326 (2003). CAS PubMed Google Scholar * Walker, F. et al. Activation of the ras/mitogen-activated protein kinase pathway by kinase-defective
epidermal growth factor receptors results in cell survival but not proliferation. _Mol. Cell Biol_. 18, 7192–7204 (1998). * Okada, T., Miyagi, H., Sako, Y., Hiroshima, M. & Mochizuki,
A. Origin of diverse phosphorylation patterns in the ERBB system. _Biophys. J_. 121, 1–11 (2021). * Giocanti, N., Hennequin, C., Rouillard, D., Defrance, R. & Favaudon, V. Additive
interaction of gefitinib (‘Iressa’, ZD1839) and ionising radiation in human tumour cells in vitro. _Br. J. Cancer_ 91, 2026–2033 (2004). Article CAS PubMed PubMed Central Google Scholar
* Sharma, A. et al. On-water NiFe2O4 nanoparticle-catalyzed one-pot synthesis of biofunctionalized pyrimidine-thiazole derivatives: In silico binding affinity and in vitro anticancer
activity studies. _ChemistrySelect_ 3, 11012–11019 (2018). Article Google Scholar * Morita, K. et al. In situ synthesis of an anticancer peptide amphiphile using tyrosine kinase
overexpressed in cancer cells. _JACS Au_ 2, 2023–2028 (2022). Article CAS PubMed PubMed Central Google Scholar * Mcswiggen, D. T. et al. A high-throughput platform for single-molecule
tracking identifies drug interaction and cellular mechanisms. _eLife_ 12, RP93183 (2024). * de Laurentiis, A., Donovan, L. & Arcaro, A. Lipid rafts and caveolae in signaling by growth
factor receptors. _Open Biochem. J._ 1, 12–32 (2007). Article PubMed PubMed Central Google Scholar * Bourseau-Guilmain, E. et al. Hypoxia regulates global membrane protein endocytosis
through caveolin-1 in cancer cells. _Nat. Commun._ 7, 11371 (2016). Article ADS CAS PubMed PubMed Central Google Scholar * Jo, U. et al. EGFR endocytosis is a novel therapeutic target
in lung cancer with wild-type EGFR. _Oncotarget_ 5, 1265–1278 (2014). Article PubMed PubMed Central Google Scholar * Heppner, D. E. & van der Vliet, A. Redox-dependent regulation of
epidermal growth factor receptor signaling. _Redox Biol._ 8, 24–27 (2016). Article CAS PubMed Google Scholar * Kim, S. Y. et al. Effects of clioquinol analogues on the hypoxia-inducible
factor pathway and intracelullar mobilization of metal ions. _Biol. Pharm. Bull._ 35, 2160–2169 (2012). Article CAS PubMed Google Scholar * Wang, Y. et al. Hypoxia promotes
ligand-independent EGF receptor signaling via hypoxia-inducible factor-mediated upregulation of caveolin-1. _Proc. Natl Acad. Sci. USA_ 109, 4892–4897 (2012). Article ADS CAS PubMed
PubMed Central Google Scholar * Wen, S. Y. et al. Doxorubicin induced ROS-dependent HIF1α activation mediates blockage of IGF1R survival signaling by IGFBP3 promotes cardiac apoptosis.
_Aging_ 15, 164–178 (2023). Article CAS PubMed PubMed Central Google Scholar * Wang, H., Jin, H. & Rapraeger, A. C. Syndecan-1 and syndecan-4 capture epidermal growth factor
receptor family members and the α3β1 integrin via binding sites in their ectodomains: novel synstatins prevent kinase capture and inhibitα6β4-integrindependent epithelial cell motility. _J.
Biol. Chem._ 290, 26103–26113 (2015). Article CAS PubMed PubMed Central Google Scholar * Alves, A. C. et al. A biophysical approach to daunorubicin interaction with model membranes:
relevance for the drug’s biological activity. _J. R. Soc. Interface_ 14, 20170408 (2017). Article PubMed PubMed Central Google Scholar * Matthews, E. E. et al. Thrombopoietin receptor
activation: transmembrane helix dimerization, rotation, and allosteric modulation. _FASEB J._ 25, 2234–2244 (2011). Article CAS PubMed PubMed Central Google Scholar * Stewart, E. L.,
Tan, S. Z., Liu, G. & Tsao, M. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR. _Transl. Lung Cancer Res._ 4, 67–81 (2015). CAS PubMed
PubMed Central Google Scholar * Maeda, R., Sato, T., Okamoto, K., Yanagawa, M. & Sako, Y. Lipid-protein interplay in dimerization of juxtamembrane domains of epidermal growth factor
receptor. _Biophys. J._ 114, 893–903 (2018). Article ADS CAS PubMed PubMed Central Google Scholar * Yoshimura, A., Longmore, G. & Lodish, H. F. Point mutation in the exoplasmic
domain of the erythropoietin receptor resulting in hormone-independent activation and tumorigenicity. _Nature_ 348, 647–649 (1990). Article ADS CAS PubMed Google Scholar * Gotoh, N.,
Tojo, A., Hino, M., Yazaki, Y. & Shibuya, M. A highly conserved tyrosine residue at codon 845 within the kinase domain is not required for the transforming activity of human epidermal
growth factor receptor. _Biochem. Biophys. Res. Commun._ 186, 768–774 (1992). Article CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We thank A. Kanayama for providing
experimental support, P. Karagiannis for editing the manuscript, and all members of the Ueda lab for discussions. The epithelial-like cell lines, CHO-K1 (RCB0285), HeLa (RCB0007), A431
(RCB0202), and the mouse pro-B cell line, Ba/F3 (RCB4476), were provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan. This project was supported by the
Translational Science and Medicine Training Program, Japan Agency for Medical Research and Development (AMED); Research Support Project for Life Science and Drug Discovery (Basis for
Supporting Innovative Drug Discovery and Life Science Research (BINDS)), AMED; Center for Supporting Drug Discovery, Life Science Research, Osaka University; and the RIKEN Program for Drug
Discovery and Medical Technology Platforms (DMP). This project is funded by AMED Grant Number JP24ama121054; AMED Grant Number JP23ym0126815 to M.U.; Japan Science and Technology Agency
(JST) CREST Grant Number JPMJCR21E1 to M.U.; Ministry of Education, Culture, Sports, Science and Technology (MEXT, Japan) Grants-in-Aid for Scientific Research(B) (18H01839) to M.H.; MEXT
Grants-in-Aid for Scientific Research(B) (22H02593) to M.H.; Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research on Innovative Areas (18H05414) to M.H.
AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Laboratory of Single Molecule Biology, Graduate School of Frontier Biosciences, Osaka University, Suita, Osaka, Japan Daisuke Watanabe, Michio
Hiroshima & Masahiro Ueda * Laboratory for Cell Signaling Dynamics, Center for Biosystems Dynamics Research, RIKEN, Suita, Osaka, Japan Daisuke Watanabe, Michio Hiroshima & Masahiro
Ueda * ZIDO Corporation, Toyonaka, Osaka, Japan Masato Yasui * Department of Biological Sciences, Graduate School of Science, Osaka University, Toyonaka, Osaka, Japan Masahiro Ueda Authors *
Daisuke Watanabe View author publications You can also search for this author inPubMed Google Scholar * Michio Hiroshima View author publications You can also search for this author
inPubMed Google Scholar * Masato Yasui View author publications You can also search for this author inPubMed Google Scholar * Masahiro Ueda View author publications You can also search for
this author inPubMed Google Scholar CONTRIBUTIONS Conceptualization: M.H. and M.U. Methodology: D.W., M.Y., and M.H. Investigation: D.W. and M.H. Funding acquisition: M.H. and M.U.
Supervision: M.U. Writing—original draft: D.W., M.H., and M.U. Writing—review & editing: M.H. and M.U. CORRESPONDING AUTHORS Correspondence to Michio Hiroshima or Masahiro Ueda. ETHICS
DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Communications_ thanks David E Heppner and the other anonymous
reviewer(s) for their contribution to the peer review of this work. A peer review file is available. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to
jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION DESCRIPTION OF ADDITIONAL SUPPLEMENTARY FILES SUPPLEMENTARY MOVIE
1 SUPPLEMENTARY MOVIE 2A SUPPLEMENTARY MOVIE 2B SUPPLEMENTARY MOVIE 3 SUPPLEMENTARY DATASET 1 SUPPLEMENTARY DATASET 2 SUPPLEMENTARY DATASET 3 REPORTING SUMMARY TRANSPARENT PEER REVIEW FILE
RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use,
sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons
licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or
other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in
the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the
copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Watanabe, D.,
Hiroshima, M., Yasui, M. _et al._ Single molecule tracking based drug screening. _Nat Commun_ 15, 8975 (2024). https://doi.org/10.1038/s41467-024-53432-w Download citation * Received: 29
December 2023 * Accepted: 08 October 2024 * Published: 17 October 2024 * DOI: https://doi.org/10.1038/s41467-024-53432-w SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative