Play all audios:
ABSTRACT The heterogeneous composition of vaccine formulations and the relatively low concentration make the characterization of the protein antigens extremely challenging.
Aluminum-containing adjuvants have been used to enhance the immune response of several antigens over the last 90 years and still remain the most commonly used. Here, we show that solid-state
NMR and isotope labeling methods can be used to characterize the structural features of the protein antigen component of vaccines and to investigate the preservation of the folding state of
proteins adsorbed on Alum hydroxide matrix, providing the way to identify the regions of the protein that are mainly affected by the presence of the inorganic matrix. l-Asparaginase from
_E. coli_ has been used as a pilot model of protein antigen. This methodology can find application in several steps of the vaccine development pipeline, from the antigen optimization,
through the design of vaccine formulation, up to stability studies and manufacturing process. SIMILAR CONTENT BEING VIEWED BY OTHERS ENGINEERING THE HYDROXYL CONTENT ON ALUMINUM OXYHYDROXIDE
NANOROD FOR ELUCIDATING THE ANTIGEN ADSORPTION BEHAVIOR Article Open access 23 June 2022 FORMALDEHYDE TREATMENT OF PROTEINS ENHANCES PROTEOLYTIC DEGRADATION BY THE ENDO-LYSOSOMAL PROTEASE
CATHEPSIN S Article Open access 14 July 2020 ENSILICATED TETANUS ANTIGEN RETAINS IMMUNOGENICITY: _IN VIVO_ STUDY AND TIME-RESOLVED SAXS CHARACTERIZATION Article Open access 08 June 2020
INTRODUCTION Vaccination is one of the major contributors to the control of infections in human population globally. Insoluble aluminum salts have been used to enhance the immunogenicity of
antigens against bacterial and viral infections since 1926, when Glenny et al. reported their use to improve the response of diphteria toxoid.1 Recently other adjuvants based on oil-in-water
emulsions (i.e. MF59, AS03) and liposomes have been used for licensed vaccines and other candidates at different stages of research and development.2,3 However, the aluminum salts are the
most commonly used adjuvants for commercial vaccines and, also due to their long-term success, they still remain the “gold standard” against a new adjuvant.4 For licensed vaccines,
aluminum(III) hydroxide (AlumOH) and aluminum(III) phosphate (AlumP) are the most commonly used adjuvants. AlumOH is a chemically crystalline aluminum(III) oxyhydroxide (AlOOH), prepared by
exposing soluble aluminum(III) salts (generally Al(H2O)6Cl3 or AlK(SO4)2) to alkaline conditions to obtain a suspension which is finally dehydrated under hydrothermal conditions. AlumP is a
noncrystalline compound generated by incorporation of phosphate which interferes with the crystallization process. AlumP can be prepared by mixing the aluminum salt Al(H2O)6Cl3 or AlK(SO4)2
with a basic solution of trisodium phosphate, or directly mixing aluminum salt with phosphate solution, followed by precipitation with sodium hydroxide. The substitution of hydroxyl groups
of AlumOH with phosphate groups results in the formation of aluminum hydroxyphosphate, Al(OH)x(PO4)y, a nonstoichiometric compound in which the ratio of hydroxyls to phosphate depends on the
precipitation conditions.4 In the final formulation the antigen is adsorbed on the Alum-based adjuvant and administered as precipitate. After the injection, a fraction of the antigen is
released in the extracellular fluid and cleared from the injection site. These adjuvants enhance the immune response by a slow release of the antigen from the injection site and, more
important, through activation of the dendritic cells and stimulation of CD4+ T cells.5 However, probably due to the complexity and several immunological pathways operating simultaneously,
the mechanism of action of aluminum adjuvants for enhancing the immune response remains not fully understood, although they have been used over many years in vaccines for human use.
Electrostatic interaction, phosphate ligand exchange, hydrogen bonding and van der Waals interactions may be involved in the adsorption mechanism depending on antigen, adjuvant, excipients,
pH and ionic strength.6 However, the interaction with the adjuvant may alter folding, conformation and stability of the antigen.6 For folded protein antigens, electrostatic forces are
reported to dominate the interaction with the AlumOH hydrated gel in a manner dependent on the pH and isoelectric point.7 Alteration of folding and native conformational state of the
epitopes may affect the immune response by influencing the antigen processing and presentation, the amount of protein bound to the adjuvant and its binding affinity.8 Also the long-term
stability of the antigen in the final formulation is extremely important for the effectiveness and commercial viability of the vaccine. Therefore, the characterization of the protein antigen
bound to the aluminum gel adjuvant is particularly relevant for the development of more effective and stable vaccine. Unfortunately, the heterogeneous composition of vaccines has hampered
for long time the biophysical and structural characterization of the protein component within the formulation. Desorption from the aluminum adjuvant and elution have often been used to
analyze post-formulation antigens. However, this strategy does not provide structural information on the antigen when adsorbed on the adjuvant. Attenuated total reflectance
Fourier-transformed infrared and fluorescence spectroscopy, circular dichroism and differential scanning calorimetry have been used to investigate adjuvant-interacting protein antigens.9,10
More recently, electron microscopy has been used to characterize the antigens in adjuvant bound states.11 However, this approach is not for general use, and it may be feasible only with
extremely large proteins or protein assemblies. In the last years, solid-state NMR is emerging as an outstanding spectroscopic technique in structural
biology,12,13,14,15,16,17,18,19,20,21,22,23,24,25 and to investigate, at atomic detail, difficult protein systems in many different states, including biosilica-entrapped proteins,
hydroxyapatite-protein composites, PEGylated and polysaccharide-conjugated proteins and proteins grafted onto nanoparticles.26,27,28,29,30,31,32,33 In particular, soluble proteins and
protein assemblies,34 membrane proteins, such as bacterial porins17 and transmembrane helix proteins,35 viral capsid components36 and RNA37 have been characterized by solid-state NMR
spectroscopy, opening up promising opportunities for the study of different classes of antigens. Here we show that atomic structural details on protein antigens adsorbed on Aluminum gel
adjuvant can be achieved by solid-state NMR from vaccine formulations obtained starting from isotopically enriched antigens and stored for several months. The following NMR analysis
performed on _Escherichia coli_ l-Asparaginase (ANSII) sheds light (i) on the folding state of the protein bound to aluminum adjuvant, (ii) on the protein regions involved in the interaction
with gel, and (iii) it provides a new tool for vaccines formulation development and stability studies. RESULTS NMR SPECTROSCOPY AlumOH adjuvanted formulation of uniformly isotopically
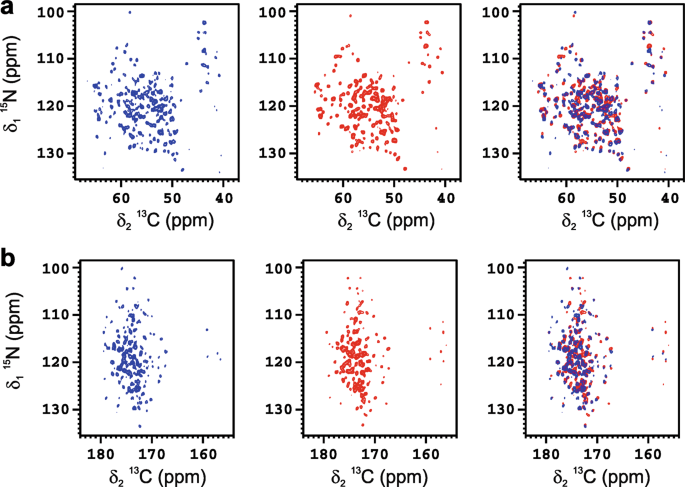
enriched ANSII [U-13C-15N] was used as vaccine model and investigated by solid-state NMR. Despite the low amount of protein absorbed on the inorganic matrix, the 2D amide-carbon alpha (2D
15N 13C NCA) and amide-carbonyl (2D 15N 13C NCO) correlation spectra of ANSII-AlumOH (Fig. 1a, b, respectively), collected on sedimented material obtained by centrifugation, are of high
quality, and comparable for the number of cross-peaks detected to the 2D 15N 13C NCA and 2D 15N 13C NCO spectra collected on rehydrated freeze-dried ANSII. In both N−C correlation spectra
the resonances are superimposable to those of the rehydrated freeze-dried ANSII (Fig. 1), immediately demonstrating that the three-dimensional structure of the protein is preserved after
adsorption on the inorganic salt. The assignment of the 2D 15N 13C NCA spectrum of ANSII-AlumOH (see Supplementary Table 1) was easily obtained by comparison with the 2D 15N 13C NCA
collected on the rehydrated freeze-dried ANSII, and also using the information from the 2D 13C-13C correlation spectrum (dipolar assisted rotational resonance, DARR) acquired on
ANSII-AlumOH. DATA ANALYSIS The assignment of the spin systems allowed the analysis of the chemical shift perturbation (CSP). The CSP data reveal that the residues experiencing the largest
changes are located on the protein surface with negative electrostatic potential (Fig. 2). In particular, the largest CSP are observed for residues in the region between Ala-250 and Lys-310,
which possesses a wide distribution of negative charge. The presence of electrostatic interactions between the negatively charged surface of the protein and the positive surface of the
inorganic material is further supported by the properties of the aluminum oxyhydroxide that has an isoelectric point of about 11.4, and at pH 7.5 exhibits a positive surface charge.38 These
findings are consistent with the mechanism proposed for the adsorption of folded antigen proteins onto the AlumOH. DISCUSSION The application of structural biology to develop new vaccines
has already proved its effectiveness.39 Engineered antigens incorporating protective determinants have been developed by NMR-based structural methodologies to improve protection, safety and
industrial scale production.40 Recently, we have demonstrated that high-resolution SSNMR, based on 13C detection, can be applied to assess the preservation of the folding of
silica-encapsulated enzymes,27 and track the chemical shift perturbation on the protein surface induced by the interaction with ligands.41 ANSII from _E. coli_ is a homotetrameric assembly
of 138 kDa with D2 symmetry. This protein is in clinical use since 1967 against childhood acute lymphoblastic leukemia but has now been largely replaced by its PEGylated form, which exhibits
longer‐lasting activity and, more important, a lower immunogenicity.42 The previously reported characterization of native ANSII and its antigenic properties make it a suitable model to
investigate the potential of new NMR methods for vaccines development and characterization. The amount of protein antigen adsorbed to the adjuvant and the heterogeneity of the vaccine
formulation are two important limiting factors for their biophysical characterization. However, these limitations can be overcome today by using the high sensitivity of solid-state NMR
combined with isotopic labeling methodologies. Therefore, the use of solid-state NMR and structural methodologies to characterize adsorbed antigens promises to solve several challenges
frequently encountered in vaccine development. For example, the destabilization of protein antigen structure upon the adsorption to the aluminum adjuvants has been suggested to play a role
in immune stimulation. In particular, protein unfolding may favor the proteolytic degradation of the antigen and the presentation of the fragments to the immune cells.6 At the same time, for
some vaccines, other studies have shown that the loss of the native secondary and tertiary structure can result in partial loss of immunogenicity.43,44 In this respect, the information
provided by solid-state NMR on folding preservation could be decisive to determine the molecular basis of the loss of efficacy and to design vaccine with an improved immunogenicity. A
further potential application of the solid-state NMR to vaccine development concerns the optimization of the experimental conditions for the adsorption of antigen to aluminum salts. Usually,
the adsorption of the antigen protein is optimized by changing the pH and buffer components that directly affect the electrostatic interactions (i.e. zeta potential). However, the amount of
protein antigen adsorbed to the aluminum salt is determined by measuring the concentration of the residual free protein in solution without any quantitative and qualitative information on
the protein bound to the adjuvant. In this respect, the collection of mono- and multidimensional solid-state NMR spectra on formulations containing isotopically enriched protein antigens
allows for a completely new approach providing semiquantitative information on the adsorbed protein antigen and on its folding state, suitable for driving structure-based optimization of
vaccine formulation and manufacturing process. Also accelerated stability studies and mechanistic studies to investigate the exposure to high temperatures, freeze-thaw events and low pH
would benefit from the use of this new methodology. Moreover, it should be pointed out that this methodology can be applied to a wide range of proteins because the labeling of antigens in
eukaryotic expression systems, although highly expensive for academia, is nowadays feasible.45,46,47,48,49,50 Finally, the extension of the cryo-probe technology to solid-state NMR and the
forthcoming increase in magnetic field strength of the NMR instruments are expected to improve further the sensitivity of the experiments allowing a more detailed characterization, or
decreasing the amount of protein required for the detection. METHODS EXPRESSION AND PURIFICATION OF UNIFORMLY ISOTOPICALLY ENRICHED ANSII [U-13C-15N] _Escherichia coli_ C41(DE3) cells were
transformed with pET-21a(+) plasmid encoding ANSII gene. The cells were cultured in 13C, 15N-enriched minimal medium (M9) containing 0.1 mg/mL of ampicillin, grown at 310 K, until OD600
reached 0.6–0.8, then induced with 1 mM isopropyl _β_-d-1-thiogalactopyranoside. They were further grown at 310 K overnight and then harvested by centrifugation at 6500 rpm (JA-10 Beckman
Coulter) for 15 min at 277 K. The pellet was suspended in 10 mM Tris-HCl, pH 8.0, 15 mM EDTA, 20% sucrose buffer (60 mL per liter of culture) and incubated at 277 K for 20 min upon magnetic
stirring. The suspension was centrifuged at 10,000 rpm (F15-6x100y Thermo Scientific) for 30 min and the supernatant discarded. The recovered pellet was re-suspended in H2O milli-Q (60 mL
per liter of culture) and newly incubated at 277 K for 20 min under magnetic stirring. Again the suspension was centrifuged at 10,000 rpm (F15-6x100y Thermo Scientific) for 30 min. The
pellet was discarded, whereas the supernatant was treated with ammonium sulfate. Still under magnetic stirring, solid ammonium sulfate was added in aliquots up to 50% saturation. The
precipitate was removed by centrifugation, then further ammonium sulfate was added up to 90% saturation to trigger the precipitation of ANSII, which was recovered again by centrifugation.
The precipitated ANSII was re-dissolved in a minimal amount of 20 mM Tris-HCl buffer at pH 8.6 and dialyzed extensively against the same buffer. ANSII was purified by anionic-exchange
chromatography using a HiPrep Q FF 16/10 column (GE Healthcare Life Science). The protein was eluted in 20 mM Tris-HCl buffer at pH 8.6 with a linear 0–1 M NaCl gradient. Fractions
containing pure ANSII were identified by Coomassie staining SDS-PAGE gels, then joined and dialyzed extensively against 50 mM phosphate buffer at pH 7.5. PREPARATION OF VACCINE FORMULATION
Commercially available AlumOH (Sigma) was used. To reduce the phosphate content and assure a complete adsorption to AlumOH adjuvant, 10 mL of ANSII [U-13C-15N] protein at the concentration
of 0.583 mg/mL in 150 mM sodium phosphate (pH 7.5) was dialyzed and concentrated to 2 mL volume by using a Vivaspin 5 kDa molecular weight cutoff membrane (Sartorius). A protein
concentration of 2.77 mg/mL was obtained as estimated by MicroBCA commercial kit (Thermo). Fifty milliliters of AlumOH adjuvanted formulation at a protein concentration of 100 µg/mL (with 2
mg/mL of Al(OH)3 and 9 mg/mL of NaCl) was prepared by mixing 1.805 mL of ANSII [U-13C-15N] (5 mg totally), 23.195 mL MilliQ H2O, 25 mL AlumOH at 4 mg/mL Alum(OH)3 and 18 mg/mL NaCl. To
estimate the amount of protein adsorbed to the AlumOH adjuvant, the hydrogel was pelleted at 15,000 rpm for 1 min and the protein content estimated on the supernatant. SAMPLE PREPARATION AND
NMR MEASUREMENTS SSNMR experiments were recorded on a Bruker Avance III spectrometer operating at 800 MHz (19 T, 201.2 MHz 13C Larmor frequency) equipped with Bruker 3.2 mm Efree NCH
probe-head. All spectra were recorded at 14 kHz MAS frequency and the sample temperature was kept at ~290 K. The sample of ANSII-AlumOH was stored for 6 months at 277 K to reproduce a
possible shortest shelf-life of a commercial vaccine. A mild centrifugation was used to separate the colloidal AlumOH adjuvant with the adsorbed ANSII from the supernatant. The hydrogel was
pelleted at 10,000 rpm for 1 h at 4 °C using an ALC multispeed refrigerated PK121R centrifuge (rotor model A-M10). Then, the precipitate was used to fill a Bruker 3.2 mm thin-wall zirconia
rotor. Silicon plugs (courtesy of Bruker Biospin) placed below the turbine cap were used to close the rotor and preserve hydration. The rotor was filled with 30 mg of wet precipitate. A
batch of freeze‐dried ANSII [U- 13C, 15N, ca. 20 mg of material] was prepared as reference sample. The protein material was packed into a Bruker 3.2 mm zirconia rotor, and rehydrated with a
solution of 9 mg/mL NaCl in MilliQ H2O, to simulate the same conditions of ANSII-AlumOH. The hydration process was monitored through 1D {1H}-13C cross-polarization SSNMR spectrum and stopped
when the resolution of the spectrum did not change any further for successive additions of the solution.26,29,30 The amount of protein present in the NMR sample of ANSII-AlumOH was
estimated to be 0.7–1 mg (per 20–25 mg of AlumOH), according to the relative intensity of 1D {1H}-13C cross-polarization spectra recorded on ANSII-AlumOH and on the sample of rehydrated
freeze-dried ANSII containing a known amount of protein. Standard 13C-detected SSNMR spectra (2D 15N 13C NCA, 2D 15N 13C NCO and 2D 13C-13C DARR, mixing time 50 ms) were acquired using the
pulse sequences reported in the literature.51 Pulses were 2.6 μs for 1H, 4 μs for 13C and 5.6 μs for 15N. The inter-scan delay was set to 1.5 s in all the experiments. The number of scans
used for the acquisition of 2D 15N 13C NCA and 15N 13C NCO experiments was 4096 and 128 for ANSII-AlumOH and rehydrated freeze-dried ANSII, respectively. Each N−C correlation experiment
collected on ANSII-AlumOH was acquired for 6 days while the 2D 13C-13C DARR (number of scans equal to 656) required 8 days of acquisition (additional experimental information is reported in
Supplementary Table 2). No significant protein desorption was observed to occur by spinning the sample at 14 kHz as proved by the comparison of the 1D {1H}-13C cross-polarization spectra
collected just after sample preparation and after the whole NMR characterization: in the two 1D {1H}-13C cross-polarization spectra the signal intensity is approximately the same
(Supplementary Fig. 1). All the spectra were processed with the Bruker TopSpin 3.2 software package and analyzed with the program CARA.52 DATA AVAILABILITY All data generated or analyzed
during this study are included in this published article and its supporting information file. REFERENCES * Glenny, A. T., Pope, C. G., Waddington, H. & Wallace, U. Immunological notes.
XVII−XXIV. _J. Pathol. Bacteriol._ 29, 31–40 (1926). Article CAS Google Scholar * Del Giudice, G., Rappuoli, R. & Didierlaurent, A. M. Correlates of adjuvanticity: a review on
adjuvants in licensed vaccines. _Semin. Immunol._ 39, 14–21 (2018). Article Google Scholar * O’Hagan, D. T., Friedland, L. R., Hanon, E. & Didierlaurent, A. M. Towards an evidence
based approach for the development of adjuvanted vaccines. _Curr. Opin. Immunol._ 47, 93–102 (2017). Article Google Scholar * HogenEsch, H., O’Hagan, D. T. & Fox, C. B. Optimizing the
utilization of aluminum adjuvants in vaccines: you might just get what you want. _NPJ Vaccin._ 3, 51 (2018). Article Google Scholar * Sokolovska, A., Hem, S. L. & HogenEsch, H.
Activation of dendritic cells and induction of CD4(+) T cell differentiation by aluminum-containing adjuvants. _Vaccine_ 25, 4575–4585 (2007). Article CAS Google Scholar * Jones, L. S. et
al. Effects of adsorption to aluminum salt adjuvants on the structure and stability of model protein antigens. _J. Biol. Chem._ 280, 13406–13414 (2005). Article CAS Google Scholar *
Jully, V., Mathot, F., Moniotte, N., Préat, V. & Lemoine, D. Mechanisms of antigen adsorption onto an aluminum-hydroxide adjuvant evaluated by high-throughput screening. _J. Pharm. Sci._
105, 1829–1836 (2016). Article CAS Google Scholar * Régnier, M. et al. Structural perturbation of diphtheria toxoid upon adsorption to aluminium hydroxide adjuvant. _Vaccine_ 30,
6783–6788 (2012). Article Google Scholar * Dong, A., Jones, L. S., Kerwin, B. A., Krishnan, S. & Carpenter, J. F. Secondary structures of proteins adsorbed onto aluminum hydroxide:
infrared spectroscopic analysis of proteins from low solution concentrations. _Anal. Biochem._ 351, 282–289 (2006). Article CAS Google Scholar * Kirkitadze, M., Sinha, A., Hu, J.,
Williams, W. & Cates, G. Adjuvanted vaccine components: analysis of structure and stability. _Procedia Vaccinol._ 1, 135–139 (2009). Article CAS Google Scholar * Soliakov, A., Harris,
J. R., Watkinson, A. & Lakey, J. H. The structure of Yersinia pestis Caf1 polymer in free and adjuvant bound states. _Vaccine_ 28, 5746–5754 (2010). Article CAS Google Scholar *
Andreas, L. B. et al. Structure of fully protonated proteins by proton-detected magic-angle spinning NMR. _Proc. Natl. Acad. Sci. USA_ 113, 9187–9192 (2016). Article CAS Google Scholar *
Quinn, C. M. et al. Dynamic regulation of HIV-1 capsid interaction with the restriction factor TRIM5α identified by magic-angle spinning NMR and molecular dynamics simulations. _Proc. Natl.
Acad. Sci. USA_ 115, 11519–11524 (2018). Article CAS Google Scholar * Page, R. C., Lee, S., Moore, J. D., Opella, S. J. & Cross, T. A. Backbone structure of a small helical integral
membrane protein: a unique structural characterization. _Protein Sci._ 18, 134–146 (2009). CAS PubMed Google Scholar * Soong, R. et al. Proton-evolved local-field solid-state NMR studies
of cytochrome b5 embedded in bicelles, revealing both structural and dynamical information. _J. Am. Chem. Soc._ 132, 5779–5788 (2010). Article CAS Google Scholar * Park, S. H. et al.
Structure of the chemokine receptor CXCR1 in phospholipid bilayers. _Nature_ 491, 779–783 (2012). Article CAS Google Scholar * Retel, J. S. et al. Structure of outer membrane protein G in
lipid bilayers. _Nat. Commun._ 8, 2073 (2017). Article Google Scholar * Fusco, G. et al. Direct observation of the three regions in α-synuclein that determine its membrane-bound
behaviour. _Nat. Commun._ 5, 3827 (2014). Article CAS Google Scholar * Sergeyev, I. V., Itin, B., Rogawski, R., Day, L. A. & McDermott, A. E. Efficient assignment and NMR analysis of
an intact virus using sequential side-chain correlations and DNP sensitization. _Proc. Natl. Acad. Sci. USA_ 114, 5171–5176 (2017). Article CAS Google Scholar * Habenstein, B. et al.
Hybrid structure of the type 1 pilus of uropathogenic _Escherichia coli_. _Angew. Chem. Int. Ed._ 54, 11691–11695 (2015). Article CAS Google Scholar * Öster, C. et al. Characterization of
protein−protein interfaces in large complexes by solid-state NMR solvent paramagnetic relaxation enhancements. _J. Am. Chem. Soc._ 139, 12165–12174 (2017). Article Google Scholar *
Tuttle, M. D. et al. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. _Nat. Struct. Mol. Biol._ 23, 409–415 (2016). Article CAS Google Scholar * Wiegand,
T. et al. The conformational changes coupling ATP hydrolysis and translocation in a bacterial DnaB helicase. _Nat. Commun._ 10, 31 (2019). Article Google Scholar * Bersch, B., Dörr, J.
M., Hessel, A., Killian, J. A. & Schanda, P. Proton-detected solid-state NMR spectroscopy of a zinc diffusion facilitator protein in native nanodiscs. _Angew. Chem. Int. Ed. Engl._ 56,
2508–2512 (2017). Article CAS Google Scholar * Joedicke, L. et al. The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors. _Nat. Chem. Biol._ 14, 284–290
(2018). Article CAS Google Scholar * Ravera, E. et al. Solid-state NMR of PEGylated proteins. _Angew. Chem. Int. Ed. Engl._ 55, 2446–2449 (2016). Article CAS Google Scholar * Fragai,
M. et al. SSNMR of biosilica-entrapped enzymes permits an easy assessment of preservation of native conformation in atomic detail. _Chem. Commun._ 50, 421–423 (2014). Article CAS Google
Scholar * Ravera, E. et al. Biosilica-entrapped enzymes studied by using dynamic nuclear-polarization-enhanced high-field NMR spectroscopy. _Chemphyschem_ 16, 2751–2754 (2015). Article CAS
Google Scholar * Giuntini, S. et al. Characterization of the conjugation pattern in large polysaccharide—protein conjugates by NMR spectroscopy. _Angew. Chem. Int. Ed._ 56, 14997–15001
(2017). Article CAS Google Scholar * Giuntini, S., Cerofolini, L., Ravera, E., Fragai, M. & Luchinat, C. Atomic structural details of a protein grafted onto gold nanoparticles. _Sci.
Rep._ 7, 17934 (2017). Article Google Scholar * Jantschke, A. et al. Insight into the supramolecular architecture of intact diatom biosilica from DNP-supported solid-state NMR
spectroscopy. _Angew. Chem. Int. Ed. Engl._ 54, 15069–15073 (2015). Article CAS Google Scholar * Martelli, T. et al. Atomic-level quality assessment of enzymes encapsulated in bioinspired
silica. _Chemistry_ 22, 425–432 (2015). Article Google Scholar * Balayssac, S. et al. Solid-state NMR of matrix metalloproteinase 12: an approach complementary to solution NMR.
_Chembiochem_ 8, 486–489 (2007). Article CAS Google Scholar * Cerofolini, L. et al. Characterization of PEGylated asparaginase: new opportunities from NMR analysis of large PEGylated
therapeutics. _Chemistry_ 25, 1984–1991 (2019). Article CAS Google Scholar * Wang, S. et al. Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane
protein. _Nat. Methods_ 10, 1007–1012 (2013). Article Google Scholar * Suiter, C. L. et al. MAS NMR of HIV-1 protein assemblies. _J. Magn. Reson._ 253, 10–22 (2015). Article CAS Google
Scholar * Marchanka, A., Simon, B., Althoff-Ospelt, G. & Carlomagno, T. RNA structure determination by solid-state NMR spectroscopy. _Nat. Commun._ 6, 7024 (2015). Article CAS Google
Scholar * Garçon, N. & Friede, M. 6—Evolution of adjuvants across the centuries. In _Plotkin’s Vaccines_ _(_ _Seventh Edition_ _)_ (eds Plotkin, S. A. et al.) 61−74.e4 (Elsevier,
Amsterdam, 2018). * Scarselli, M. et al. Rational design of a meningococcal antigen inducing broad protective immunity. _Sci. Transl. Med._ 3, 91ra62 (2011). Article CAS Google Scholar *
Cozzi, R., Scarselli, M. & Ferlenghi, I. Structural vaccinology: a three-dimensional view for vaccine development. _Curr. Top. Med. Chem._ 13, 2629–2637 (2013). Article Google Scholar
* Cerofolini, L. et al. High-resolution solid-state NMR characterization of ligand binding to a protein immobilized in a silica matrix. _J. Phys. Chem. B_ 121, 8094–8101 (2017). Article CAS
Google Scholar * Graham, M. L. Pegaspargase: a review of clinical studies. _Adv. Drug Deliv. Rev._ 55, 1293–1302 (2003). Article CAS Google Scholar * Kumru, O. S. et al. Vaccine
instability in the cold chain: mechanisms, analysis and formulation strategies. _Biologicals_ 42, 237–259 (2014). Article CAS Google Scholar * Braun, L. J. et al. Characterization of a
thermostable hepatitis B vaccine formulation. _Vaccine_ 27, 4609–4614 (2009). Article Google Scholar * Sastry, M., Bewley, C. A. & Kwong, P. D. Mammalian expression of isotopically
labeled proteins for NMR spectroscopy. _Adv. Exp. Med. Biol._ 992, 197–211 (2012). Article CAS Google Scholar * Werner, K., Richter, C., Klein-Seetharaman, J. & Schwalbe, H. Isotope
labeling of mammalian GPCRs in HEK293 cells and characterization of the C-terminus of bovine rhodopsin by high resolution liquid NMR spectroscopy. _J. Biomol. NMR_ 40, 49–53 (2008). Article
CAS Google Scholar * Vajpai, N. et al. Solution conformations and dynamics of ABL kinase-inhibitor complexes determined by NMR substantiate the different binding modes of
imatinib/nilotinib and dasatinib. _J. Biol. Chem._ 283, 18292–18302 (2008). Article CAS Google Scholar * Strauss, A. et al. Efficient uniform isotope labeling of Abl kinase expressed in
Baculovirus-infected insect cells. _J. Biomol. NMR_ 31, 343–349 (2005). Article CAS Google Scholar * Morgan, W. D., Kragt, A. & Feeney, J. Expression of deuterium-isotope-labelled
protein in the yeast pichia pastoris for NMR studies. _J. Biomol. NMR_ 17, 337–347 (2000). Article CAS Google Scholar * Takahashi, H. & Shimada, I. Production of isotopically labeled
heterologous proteins in non-E. coli prokaryotic and eukaryotic cells. _J. Biomol. NMR_ 46, 3–10 (2010). Article CAS Google Scholar * Schuetz, A. et al. Protocols for the sequential
solid-state NMR spectroscopic assignment of a uniformly labeled 25 kDa protein: HET-s(1-227). _ChemBioChem_ 11, 1543–1551 (2010). Article CAS Google Scholar * Keller, R. _The Computer
Aided Resonance Assignment Tutorial (CARA)_ (CANTINA Verlag, Goldau, 2004). * Grzesiek, S. et al. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of
the binding surface for the SH3 domain of Hck tyrosine protein kinase. _Nat. Struct. Biol._ 3, 340–345 (1996). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS This work
has been supported by Regione Toscana (CERM-TT and BioEnable), Fondazione Cassa di Risparmio di Firenze and COST Action CA15209 “European Network on NMR Relaxometry”. The authors acknowledge
the support and the use of resources of Instruct-ERIC, a landmark ESFRI project, and specifically the CERM/CIRMMP Italy center. The authors acknowledge the support of the University of
Florence and the _Recombinant Proteins JOYNLAB_. The FP7 WeNMR (project no. 261572) and H2020 West-Life (project no. 675858) European e-Infrastructure projects are acknowledged for the use
of their web portals. These portals make use of the EGI infrastructure and DIRAC4EGI service with the dedicated support of CESNET-MetaCloud, INFN-PADOVA, NCG-INGRID-PT, RAL-LCG2, TW-NCHC,
IFCA-LCG2, SURFsara and NIKHEF, and the additional support of the national GRID Initiatives of Belgium, France, Italy, Germany, the Netherlands, Poland, Portugal, Spain, UK, South Africa,
Malaysia, Taiwan and the US Open Science Grid. Funding from The Italian Ministry of Education, Universities and Research (MIUR) PRIN project no. 2012SK7ASN and the EC Contracts iNext no.
653706 is acknowledged. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Magnetic Resonance Center (CERM), University of Florence and Consorzio Interuniversitario Risonanze Magnetiche di
Metallo Proteine (CIRMMP), Via L. Sacconi 6, 50019, Sesto Fiorentino, Italy Linda Cerofolini, Enrico Ravera, Claudio Luchinat & Marco Fragai * Department of Chemistry, University of
Florence, Via della Lastruccia 3, 50019, Sesto Fiorentino, Italy Stefano Giuntini, Enrico Ravera, Claudio Luchinat & Marco Fragai * Technical R&D, GSK Vaccines, Via Fiorentina 1,
53100, Siena, Italy Francesco Berti Authors * Linda Cerofolini View author publications You can also search for this author inPubMed Google Scholar * Stefano Giuntini View author
publications You can also search for this author inPubMed Google Scholar * Enrico Ravera View author publications You can also search for this author inPubMed Google Scholar * Claudio
Luchinat View author publications You can also search for this author inPubMed Google Scholar * Francesco Berti View author publications You can also search for this author inPubMed Google
Scholar * Marco Fragai View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.F., F.B. and C.L. designed the research; S.G. expressed and
purified the protein and prepared the formulation; L.C., M.F., E.R., and S.G. performed the NMR experiments; L.C. performed the resonance assignment; L.C., M.F., E.R. and C.L. performed the
data analysis; all authors wrote the manuscript, read and approved the completed version. CORRESPONDING AUTHORS Correspondence to Francesco Berti or Marco Fragai. ETHICS DECLARATIONS
COMPETING INTERESTS F.B. is an employee of the GSK group of companies. All other authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE: Springer Nature remains
neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTAL MATERIAL RIGHTS AND PERMISSIONS OPEN ACCESS This article
is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you
give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material
in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative
Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a
copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Cerofolini, L., Giuntini, S., Ravera, E. _et al._
Structural characterization of a protein adsorbed on aluminum hydroxide adjuvant in vaccine formulation. _npj Vaccines_ 4, 20 (2019). https://doi.org/10.1038/s41541-019-0115-7 Download
citation * Received: 12 January 2019 * Accepted: 03 May 2019 * Published: 28 May 2019 * DOI: https://doi.org/10.1038/s41541-019-0115-7 SHARE THIS ARTICLE Anyone you share the following link
with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt
content-sharing initiative