Play all audios:
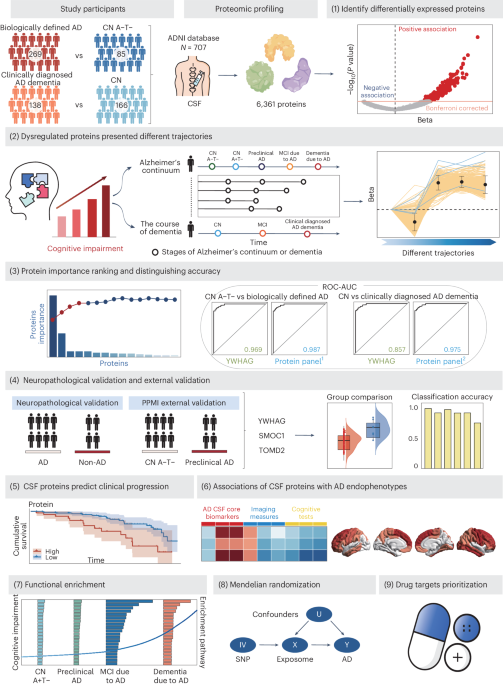
ABSTRACT Recent expansion of proteomic coverage opens unparalleled avenues to unveil new biomarkers of Alzheimer’s disease (AD). Among 6,361 cerebrospinal fluid (CSF) proteins analysed from
the ADNI database, YWHAG performed best in diagnosing both biologically (AUC = 0.969) and clinically (AUC = 0.857) defined AD. Four- (YWHAG, SMOC1, PIGR and TMOD2) and five- (ACHE, YWHAG,
PCSK1, MMP10 and IRF1) protein panels greatly improved the accuracy to 0.987 and 0.975, respectively. Their superior performance was validated in an independent external cohort and in
discriminating autopsy-confirmed AD versus non-AD, rivalling even canonical CSF ATN biomarkers. Moreover, they effectively predicted the clinical progression to AD dementia and were strongly
associated with AD core biomarkers and cognitive decline. Synaptic, neurogenic and infectious pathways were enriched in distinct AD stages. Mendelian randomization did not support the
significant genetic link between CSF proteins and AD. Our findings revealed promising high-performance biomarkers for AD diagnosis and prediction, with implications for clinical trials
targeting different pathomechanisms. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through
your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this
journal Receive 12 digital issues and online access to articles $119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article
PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact
customer support SIMILAR CONTENT BEING VIEWED BY OTHERS CSF PROTEOME PROFILING REVEALS BIOMARKERS TO DISCRIMINATE DEMENTIA WITH LEWY BODIES FROM ALZHEIMER´S DISEASE Article Open access 13
September 2023 CSF PROTEOME PROFILING ACROSS THE ALZHEIMER’S DISEASE SPECTRUM REFLECTS THE MULTIFACTORIAL NATURE OF THE DISEASE AND IDENTIFIES SPECIFIC BIOMARKER PANELS Article 10 November
2022 CEREBROSPINAL FLUID PROTEOMICS IN PATIENTS WITH ALZHEIMER’S DISEASE REVEALS FIVE MOLECULAR SUBTYPES WITH DISTINCT GENETIC RISK PROFILES Article Open access 09 January 2024 DATA
AVAILABILITY Data used in the preparation of this Article were obtained on 12 September 2023 from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database
(https://ida.loni.usc.edu/pages/access/studyData.jsp?project=ADNI) and the Parkinson’s Progression Markers Initiative (PPMI) database (www.ppmi-info.org/access-data-specimens/download-data)
(RRID: SCR_006431). For up-to-date information on the study, visit http://adni.loni.usc.edu/ and www.ppmi-info.org. ADNI data are publicly available to bona fide researchers upon application
at https://adni.loni.usc.edu/, and PPMI data are publicly available to bona fide researchers upon application at https://www.ppmi-info.org/. Enrichment analysis data can be obtained from
the STRING website (https://cn.string-db.org/). Agora Druggability data can be obtained from https://www.synapse.org/. All data supporting the findings described in this paper are available
within the paper, in the Supplementary Information and from the corresponding author upon request. Source data are provided with this paper. CODE AVAILABILITY All software used in this study
is publicly available. The code used in this study can be accessed via GitHub at https://github.com/jasonHKU0907/AD_CSF_ADNI ref. 75. REFERENCES * Johnson, E. C. B. et al. Large-scale deep
multi-layer analysis of Alzheimer’s disease brain reveals strong proteomic disease-related changes not observed at the RNA level. _Nat. Neurosci._ 25, 213–225 (2022). CAS PubMed PubMed
Central Google Scholar * van der Kant, R., Goldstein, L. S. B. & Ossenkoppele, R. Amyloid-beta-independent regulators of tau pathology in Alzheimer disease. _Nat. Rev. Neurosci._ 21,
21–35 (2020). PubMed Google Scholar * Sung, Y. J. et al. Proteomics of brain, CSF, and plasma identifies molecular signatures for distinguishing sporadic and genetic Alzheimer’s disease.
_Sci. Transl. Med._ 15, eabq5923 (2023). CAS PubMed PubMed Central Google Scholar * Johnson, E. C. B. et al. Cerebrospinal fluid proteomics define the natural history of autosomal
dominant Alzheimer’s disease. _Nat. Med._ 29, 1979–1988 (2023). CAS PubMed PubMed Central Google Scholar * Del Campo, M. et al. CSF proteome profiling across the Alzheimer’s disease
spectrum reflects the multifactorial nature of the disease and identifies specific biomarker panels. _Nat. Aging_ 2, 1040–1053 (2022). PubMed PubMed Central Google Scholar * Haque, R. et
al. A protein panel in cerebrospinal fluid for diagnostic and predictive assessment of Alzheimer’s disease. _Sci. Transl. Med._ 15, eadg4122 (2023). CAS PubMed PubMed Central Google
Scholar * Libiger, O. et al. Longitudinal CSF proteomics identifies NPTX2 as a prognostic biomarker of Alzheimer’s disease. _Alzheimers Dement._ 17, 1976–1987 (2021). CAS PubMed Google
Scholar * Wildsmith, K. R. et al. Identification of longitudinally dynamic biomarkers in Alzheimer’s disease cerebrospinal fluid by targeted proteomics. _Mol. Neurodegener._ 9, 22 (2014).
PubMed PubMed Central Google Scholar * Bader, J. M. et al. Proteome profiling in cerebrospinal fluid reveals novel biomarkers of Alzheimer’s disease. _Mol. Syst. Biol._ 16, e9356 (2020).
CAS PubMed PubMed Central Google Scholar * Whelan, C. D. et al. Multiplex proteomics identifies novel CSF and plasma biomarkers of early Alzheimer’s disease. _Acta Neuropathol. Commun._
7, 169 (2019). PubMed PubMed Central Google Scholar * Modeste, E. S. et al. Quantitative proteomics of cerebrospinal fluid from African Americans and Caucasians reveals shared and
divergent changes in Alzheimer’s disease. _Mol. Neurodegener._ 18, 48 (2023). CAS PubMed PubMed Central Google Scholar * Jack, C. R. Jr. et al. NIA-AA Research Framework: toward a
biological definition of Alzheimer’s disease. _Alzheimers Dement._ 14, 535–562 (2018). PubMed Google Scholar * Jiang, T. et al. TREM1 facilitates microglial phagocytosis of amyloid beta.
_Acta Neuropathol._ 132, 667–683 (2016). CAS PubMed Google Scholar * Replogle, J. M. et al. A TREM1 variant alters the accumulation of Alzheimer-related amyloid pathology. _Ann. Neurol._
77, 469–477 (2015). CAS PubMed PubMed Central Google Scholar * Lomoio, S. et al. Gga3 deletion and a GGA3 rare variant associated with late onset Alzheimer’s disease trigger BACE1
accumulation in axonal swellings. _Sci. Transl. Med._ 12, eaba1871 (2020). CAS PubMed PubMed Central Google Scholar * Bai, B. et al. Deep multilayer brain proteomics identifies molecular
networks in Alzheimer’s disease progression. _Neuron_ 105, 975–991.e7 (2020). CAS PubMed PubMed Central Google Scholar * Hong, H. et al. Cross-talking pathways of rapidly accelerated
fibrosarcoma-1 (RAF-1) in Alzheimer’s disease. _Mol. Neurobiol_. 61, 2798–2807 (2024). * Park, H., Lee, Y. B. & Chang, K. A. miR-200c suppression increases tau hyperphosphorylation by
targeting 14-3-3γ in early stage of 5xFAD mouse model of Alzheimer’s disease. _Int. J. Biol. Sci._ 18, 2220–2234 (2022). CAS PubMed PubMed Central Google Scholar * Higginbotham, L. et
al. Integrated proteomics reveals brain-based cerebrospinal fluid biomarkers in asymptomatic and symptomatic Alzheimer’s disease. _Sci. Adv._ 6, eaaz9360 (2020). CAS PubMed PubMed Central
Google Scholar * Zhou, M. et al. Targeted mass spectrometry to quantify brain-derived cerebrospinal fluid biomarkers in Alzheimer’s disease. _Clin. Proteom._ 17, 19 (2020). Google Scholar
* Roy, D. S. et al. Anterior thalamic dysfunction underlies cognitive deficits in a subset of neuropsychiatric disease models. _Neuron_ 109, 2590–2603.e13 (2021). CAS PubMed PubMed
Central Google Scholar * Johnson, E. C. B. et al. Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated
with microglia and astrocyte activation. _Nat. Med._ 26, 769–780 (2020). CAS PubMed PubMed Central Google Scholar * Watson, C. M. et al. Quantitative mass spectrometry analysis of
cerebrospinal fluid protein biomarkers in Alzheimer’s disease. _Sci. Data_ 10, 261 (2023). CAS PubMed PubMed Central Google Scholar * Dammer, E. B. et al. Multi-platform proteomic
analysis of Alzheimer’s disease cerebrospinal fluid and plasma reveals network biomarkers associated with proteostasis and the matrisome. _Alzheimers Res. Ther._ 14, 174 (2022). CAS PubMed
PubMed Central Google Scholar * van der Ende, E. L. et al. CSF proteomics in autosomal dominant Alzheimer’s disease highlights parallels with sporadic disease. _Brain_ 146, 4495–4507
(2023). PubMed PubMed Central Google Scholar * Scheltens, P. et al. Alzheimer’s disease. _Lancet_ 397, 1577–1590 (2021). CAS PubMed PubMed Central Google Scholar * Omotade, O. F. et
al. Tropomodulin isoform-specific regulation of dendrite development and synapse formation. _J. Neurosci._ 38, 10271–10285 (2018). CAS PubMed PubMed Central Google Scholar * Folon, L. et
al. Contribution of heterozygous PCSK1 variants to obesity and implications for precision medicine: a case–control study. _Lancet Diabetes Endocrinol._ 11, 182–190 (2023). CAS PubMed
Google Scholar * Deng, Y. T. et al. Association of life course adiposity with risk of incident dementia: a prospective cohort study of 322,336 participants. _Mol. Psychiatry_ 27, 3385–3395
(2022). CAS PubMed Google Scholar * Zhang, J. R. et al. The polymeric immunoglobulin receptor translocates pneumococci across human nasopharyngeal epithelial cells. _Cell_ 102, 827–837
(2000). CAS PubMed Google Scholar * Rosain, J. et al. Human IRF1 governs macrophagic IFN-γ immunity to mycobacteria. _Cell_ 186, 621–645.e33 (2023). CAS PubMed PubMed Central Google
Scholar * Iovino, F. et al. pIgR and PECAM-1 bind to pneumococcal adhesins RrgA and PspC mediating bacterial brain invasion. _J. Exp. Med._ 214, 1619–1630 (2017). CAS PubMed PubMed
Central Google Scholar * Sudwarts, A. et al. BIN1 is a key regulator of proinflammatory and neurodegeneration-related activation in microglia. _Mol. Neurodegener._ 17, 33 (2022). CAS
PubMed PubMed Central Google Scholar * Morris, J. C. et al. Autosomal dominant and sporadic late onset Alzheimer’s disease share a common in vivo pathophysiology. _Brain_ 145, 3594–3607
(2022). PubMed PubMed Central Google Scholar * Del Campo, M. et al. CSF proteome profiling reveals biomarkers to discriminate dementia with Lewy bodies from Alzheimer’s disease. _Nat.
Commun._ 14, 5635 (2023). PubMed PubMed Central Google Scholar * Timsina, J. et al. Comparative analysis of Alzheimer’s disease cerebrospinal fluid biomarkers measurement by multiplex
SOMAscan platform and immunoassay-based approach. _J. Alzheimers Dis._ 89, 193–207 (2022). CAS PubMed PubMed Central Google Scholar * Rutledge, J. et al. Comprehensive proteomics of CSF,
plasma, and urine identify DDC and other biomarkers of early Parkinson’s disease. _Acta Neuropathol._ 147, 52 (2024). CAS PubMed PubMed Central Google Scholar * Dammer, E. B. et al.
Proteomic analysis of Alzheimer's disease cerebrospinal fluid reveals alterations associated with APOE epsilon4 and atomoxetine treatment. _Sci. Transl. Med._ 16, eadn3504 (2024). CAS
PubMed Google Scholar * Eldjarn, G. H. et al. Large-scale plasma proteomics comparisons through genetics and disease associations. _Nature_ 622, 348–358 (2023). CAS PubMed PubMed Central
Google Scholar * Barranco, N. et al. Dense core vesicle markers in CSF and cortical tissues of patients with Alzheimer’s disease. _Transl. Neurodegener._ 10, 37 (2021). CAS PubMed
PubMed Central Google Scholar * Garcia-Ayllon, M. S., Silveyra, M. X. & Saez-Valero, J. Association between acetylcholinesterase and beta-amyloid peptide in Alzheimer’s cerebrospinal
fluid. _Chem. Biol. Interact._ 175, 209–215 (2008). CAS PubMed Google Scholar * Montoliu-Gaya, L. et al. Mass spectrometric simultaneous quantification of tau species in plasma shows
differential associations with amyloid and tau pathologies. _Nat. Aging_ 3, 661–669 (2023). CAS PubMed PubMed Central Google Scholar * Barthélemy, N. R. et al. CSF tau phosphorylation
occupancies at T217 and T205 represent improved biomarkers of amyloid and tau pathology in Alzheimer’s disease. _Nat. Aging_ 3, 391–401 (2023). PubMed PubMed Central Google Scholar *
Ossenkoppele, R. et al. Amyloid and tau PET-positive cognitively unimpaired individuals are at high risk for future cognitive decline. _Nat. Med._ 28, 2381–2387 (2022). CAS PubMed PubMed
Central Google Scholar * Yang, C. et al. Genomic atlas of the proteome from brain, CSF and plasma prioritizes proteins implicated in neurological disorders. _Nat. Neurosci._ 24, 1302–1312
(2021). CAS PubMed PubMed Central Google Scholar * Gold, L. et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. _PLoS ONE_ 5, e15004 (2010). CAS PubMed
PubMed Central Google Scholar * Candia, J. et al. Assessment of variability in the SOMAscan assay. _Sci. Rep._ 7, 14248 (2017). PubMed PubMed Central Google Scholar * Bittner, T. et al.
Technical performance of a novel, fully automated electrochemiluminescence immunoassay for the quantitation of beta-amyloid (1-42) in human cerebrospinal fluid. _Alzheimers Dement._ 12,
517–526 (2016). PubMed Google Scholar * Irwin, D. J. et al. Evolution of Alzheimer’s disease cerebrospinal fluid biomarkers in early Parkinson’s disease. _Ann. Neurol._ 88, 574–587 (2020).
CAS PubMed PubMed Central Google Scholar * Schindler, S. E. et al. Cerebrospinal fluid biomarkers measured by Elecsys assays compared to amyloid imaging. _Alzheimers Dement._ 14,
1460–1469 (2018). PubMed Google Scholar * Meyer, P. F., Pichet Binette, A., Gonneaud, J., Breitner, J. C. S. & Villeneuve, S. Characterization of Alzheimer disease biomarker
discrepancies using cerebrospinal fluid phosphorylated tau and AV1451 positron emission tomography. _JAMA Neurol._ 77, 508–516 (2020). PubMed PubMed Central Google Scholar * Weinshel, S.
et al. Appropriateness of applying cerebrospinal fluid biomarker cutoffs from Alzheimer’s disease to Parkinson’s disease. _J. Parkinsons Dis._ 12, 1155–1167 (2022). CAS PubMed PubMed
Central Google Scholar * Franklin, E. E. et al. Brain collection, standardized neuropathologic assessment, and comorbidity in Alzheimer’s Disease Neuroimaging Initiative 2 participants.
_Alzheimers Dement._ 11, 815–822 (2015). PubMed Google Scholar * Jack, C. R. Jr. et al. Different definitions of neurodegeneration produce similar amyloid/neurodegeneration biomarker group
findings. _Brain_ 138, 3747–3759 (2015). PubMed PubMed Central Google Scholar * Palmqvist, S., Mattsson, N., Hansson, O. & Alzheimer’s Disease Neuroimaging Initiative. Cerebrospinal
fluid analysis detects cerebral amyloid-beta accumulation earlier than positron emission tomography. _Brain_ 139, 1226–1236 (2016). PubMed PubMed Central Google Scholar * Desikan, R. S.
et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. _Neuroimage_ 31, 968–980 (2006). PubMed Google Scholar *
Gibbons, L. E. et al. A composite score for executive functioning, validated in Alzheimer’s Disease Neuroimaging Initiative (ADNI) participants with baseline mild cognitive impairment.
_Brain Imaging Behav._ 6, 517–527 (2012). PubMed PubMed Central Google Scholar * Crane, P. K. et al. Development and assessment of a composite score for memory in the Alzheimer’s Disease
Neuroimaging Initiative (ADNI). _Brain Imaging Behav._ 6, 502–516 (2012). PubMed PubMed Central Google Scholar * Donohue, M. C. et al. The preclinical Alzheimer cognitive composite:
measuring amyloid-related decline. _JAMA Neurol._ 71, 961–970 (2014). PubMed PubMed Central Google Scholar * Wang, Y. et al. Phosphorylated α-synuclein in Parkinson’s disease. _Sci.
Transl. Med._ 4, 121ra120 (2012). Google Scholar * Guolin, K. et al. LightGBM: a highly efficient gradient boosting decision tree. _Adv. Neural Inf. Process. Syst._ 30, 3149–3157 (2017).
Google Scholar * DeLong, E. R., DeLong, D. M. & Clarke-Pearson, D. L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric
approach. _Biometrics_ 44, 837–845 (1988). CAS PubMed Google Scholar * Delaneau, O., Zagury, J. F. & Marchini, J. Improved whole-chromosome phasing for disease and population genetic
studies. _Nat. Methods_ 10, 5–6 (2013). CAS PubMed Google Scholar * Das, S. et al. Next-generation genotype imputation service and methods. _Nat. Genet._ 48, 1284–1287 (2016). CAS PubMed
PubMed Central Google Scholar * Zheng, J. et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. _Nat. Genet._ 52, 1122–1131
(2020). CAS PubMed PubMed Central Google Scholar * Pietzner, M. et al. Mapping the proteo-genomic convergence of human diseases. _Science_ 374, eabj1541 (2021). PubMed PubMed Central
Google Scholar * Ferkingstad, E. et al. Large-scale integration of the plasma proteome with genetics and disease. _Nat. Genet._ 53, 1712–1721 (2021). CAS PubMed Google Scholar * Hansson,
O. et al. The genetic regulation of protein expression in cerebrospinal fluid. _EMBO Mol. Med._ 15, e16359 (2023). CAS PubMed Google Scholar * Kunkle, B. W. et al. Genetic meta-analysis
of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. _Nat. Genet._ 51, 414–430 (2019). CAS PubMed PubMed Central Google Scholar
* Burgess, S., Davies, N. M. & Thompson, S. G. Bias due to participant overlap in two-sample Mendelian randomization. _Genet. Epidemiol._ 40, 597–608 (2016). PubMed PubMed Central
Google Scholar * Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. _Genet. Epidemiol._ 37, 658–665
(2013). PubMed PubMed Central Google Scholar * Bowden, J. et al. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the
role of the I2 statistic. _Int. J. Epidemiol._ 45, 1961–1974 (2016). PubMed PubMed Central Google Scholar * Burgess, S. & Thompson, S. G. Avoiding bias from weak instruments in
Mendelian randomization studies. _Int. J. Epidemiol._ 40, 755–764 (2011). PubMed Google Scholar * Burgess, S. Sample size and power calculations in Mendelian randomization with a single
instrumental variable and a binary outcome. _Int. J. Epidemiol._ 43, 922–929 (2014). PubMed PubMed Central Google Scholar * You, J. et al. jasonHKU0907 / AD_CSF_ADNI. _GitHub_
https://github.com/jasonHKU0907/AD_CSF_ADNI (2024). Download references ACKNOWLEDGEMENTS We thank all participants who donated their brains to the ADNI Neuropathology Core Center and PPMI
database. We also thank all investigators who collected and processed specimens and performed neuropathological assessments in ADNI and PPMI. As such, the investigators within the ADNI or
PPMI contributed to the design and implementation of the database and/or provided data but did not participate in the analysis or in the writing of this paper. A complete listing of ADNI
investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf. We also thank all contributors to the ADNI and PPMI databases. Data
collection and sharing for this project was funded by the ADNI (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH12-2-0012). ADNI is
funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s
Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai, Inc.; Elan Pharmaceuticals,
Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research
& Development, LLC; Johnson & Johnson Pharmaceutical Research & Development LLC; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC; NeuroRx Research;
Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer, Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of
Health Research provides funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org).
The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of
Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. PPMI, a public–private partnership, is funded by the Michael J.
Fox Foundation for Parkinson’s Research and funding partners, including 4D Pharma, Abbvie, AcureX, Allergan, Amathus Therapeutics, Aligning Science Across Parkinson’s, AskBio, Avid
Radiopharmaceuticals, BIAL, BioArctic, Biogen, Biohaven, BioLegend, BlueRock Therapeutics, Bristol Myers Squibb, Calico Labs, Capsida Biotherapeutics, Celgene, Cerevel Therapeutics, Coave
Therapeutics, DaCapo Brainscience, Denali, Edmond J. Safra Foundation, Eli Lilly, Gain Therapeutics, GE HealthCare, Genentech, GSK, Golub Capital, Handl Therapeutics, Insitro, Jazz
Pharmaceuticals, Johnson & Johnson Innovative Medicine, Lundbeck, Merck, Meso Scale Discovery, Mission Therapeutics, Neurocrine Biosciences, Neuron23, Neuropore, Pfizer, Piramal, Prevail
Therapeutics, Roche, Sanofi, Servier, Sun Pharma Advanced Research Company, Takeda, Teva, UCB, Vanqua Bio, Verily, Voyager Therapeutics, the Weston Family Foundation and Yumanity
Therapeutics. We also thank all the participants and researchers from Agora (10.57718/agora-adknowledgeportal), a platform initially developed by the NIA-funded Accelerating Medicines
Partnership in AD consortium that shares evidence in support of AD target discovery. J.T.-Y. was funded by grants from the Science and Technology Innovation 2030 Major Projects
(2022ZD0211600), the National Natural Science Foundation of China (92249305), the Shanghai Municipal Science and Technology Major Project (2023SHZDZX02) and the Shanghai Municipal Health
Commission Emerging Interdisciplinary Research Project (2022JC01). W.C. was funded by the National Natural Science Foundation of China (82071997) and the Shanghai Rising-Star Program
(21QA1408700). J.F.-F. was funded by the National Key R&D Program of China (2018YFC1312904, 2019YFA0709502), the Shanghai Municipal Science and Technology Major Project (2018SHZDZX01)
and the 111 Project (No. B18015). Y.G. was funded by the National Postdoctoral Program for Innovative Talents (BX20240073). J.Y. was funded by the Shanghai Pujiang Talent Program (23PJD006).
S.D.-C. was funded by the National Postdoctoral Program for Innovative Talents (BX20230087). Further, we thank the ZHANGJIANG LAB, Tianqiao and Chrissy Chen Institute, and the State Key
Laboratory of Neurobiology and Frontiers Center for Brain Science of the Ministry of Education, Fudan University for support. The funders had no role in study design, data collection and
analysis, decision to publish or preparation of the manuscript. AUTHOR INFORMATION Author notes * These authors contributed equally: Yu Guo, Shi-Dong Chen, Jia You, Shu-Yi Huang, Yi-Lin
Chen. AUTHORS AND AFFILIATIONS * Department of Neurology and National Center for Neurological Disorders, Huashan Hospital, State Key Laboratory of Medical Neurobiology and MOE Frontiers
Center for Brain Science, Shanghai Medical College, Fudan University, Shanghai, China Yu Guo, Shi-Dong Chen, Jia You, Shu-Yi Huang, Yi-Lin Chen, Yi Zhang, Xiao-Yu He, Yue-Ting Deng, Ya-Ru
Zhang, Yu-Yuan Huang, Qiang Dong, Wei Cheng & Jin-Tai Yu * Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China Jia You, Lin-Bo Wang,
Jian-Feng Feng & Wei Cheng * Key Laboratory of Computational Neuroscience and Brain-Inspired Intelligence, Fudan University, Ministry of Education, Shanghai, China Jian-Feng Feng &
Wei Cheng Authors * Yu Guo View author publications You can also search for this author inPubMed Google Scholar * Shi-Dong Chen View author publications You can also search for this author
inPubMed Google Scholar * Jia You View author publications You can also search for this author inPubMed Google Scholar * Shu-Yi Huang View author publications You can also search for this
author inPubMed Google Scholar * Yi-Lin Chen View author publications You can also search for this author inPubMed Google Scholar * Yi Zhang View author publications You can also search for
this author inPubMed Google Scholar * Lin-Bo Wang View author publications You can also search for this author inPubMed Google Scholar * Xiao-Yu He View author publications You can also
search for this author inPubMed Google Scholar * Yue-Ting Deng View author publications You can also search for this author inPubMed Google Scholar * Ya-Ru Zhang View author publications You
can also search for this author inPubMed Google Scholar * Yu-Yuan Huang View author publications You can also search for this author inPubMed Google Scholar * Qiang Dong View author
publications You can also search for this author inPubMed Google Scholar * Jian-Feng Feng View author publications You can also search for this author inPubMed Google Scholar * Wei Cheng
View author publications You can also search for this author inPubMed Google Scholar * Jin-Tai Yu View author publications You can also search for this author inPubMed Google Scholar
CONTRIBUTIONS J.-T.Y. conceptualized and designed the study, interpreted the data and revised the manuscript. Y.G., S.-D.C., J.Y., S.-Y.H. and Y.-L.C. collected, analysed and interpreted the
data, and drafted and revised the manuscript. All authors participated in the revision of the manuscript, had full access to all the study data and accept responsibility for submission of
the paper for publication. CORRESPONDING AUTHOR Correspondence to Jin-Tai Yu. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW
INFORMATION _Nature Human Behaviour_ thanks Gajanan Sathe, Christopher Whelan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. ADDITIONAL
INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY
INFORMATION Supplementary Figs. 1–4. REPORTING SUMMARY SUPPLEMENTARY TABLES Supplementary Tables 1–21. SOURCE DATA FOR SUPPLEMENTARY FIGURES Source Data for Supplementary Figs. 1–4. SOURCE
DATA SOURCE DATA FIG. 1 Data to plot Fig. 1. SOURCE DATA FIG. 2 Data to plot Fig. 2. SOURCE DATA FIG. 3 Data to plot Fig. 3. SOURCE DATA FIG. 4 Data to plot Fig. 4. SOURCE DATA FIG. 5 Data
to plot Fig. 5. SOURCE DATA FIG. 6 Data to plot Fig. 6. RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under
a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such
publishing agreement and applicable law. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Guo, Y., Chen, SD., You, J. _et al._ Multiplex cerebrospinal fluid proteomics
identifies biomarkers for diagnosis and prediction of Alzheimer’s disease. _Nat Hum Behav_ 8, 2047–2066 (2024). https://doi.org/10.1038/s41562-024-01924-6 Download citation * Received: 16
November 2023 * Accepted: 10 June 2024 * Published: 10 July 2024 * Issue Date: October 2024 * DOI: https://doi.org/10.1038/s41562-024-01924-6 SHARE THIS ARTICLE Anyone you share the
following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer
Nature SharedIt content-sharing initiative