Play all audios:
ABSTRACT LY6E is an antiviral restriction factor that inhibits coronavirus spike-mediated fusion, but the cell types in vivo that require LY6E for protection from respiratory coronavirus
infection are unknown. Here we used a panel of seven conditional _Ly6e_ knockout mice to define which _Ly6e_-expressing cells confer control of airway infection by murine coronavirus and
severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Loss of _Ly6e_ in _Lyz2_-expressing cells, radioresistant _Vav1_-expressing cells and non-haematopoietic cells increased
susceptibility to murine coronavirus. Global conditional loss of _Ly6e_ expression resulted in clinical disease and higher viral burden after SARS-CoV-2 infection, but little evidence of
immunopathology. We show that _Ly6e_ expression protected secretory club and ciliated cells from SARS-CoV-2 infection and prevented virus-induced loss of an epithelial cell transcriptomic
signature in the lung. Our study demonstrates that lineage confined rather than broad expression of _Ly6e_ sufficiently confers resistance to disease caused by murine and human
coronaviruses. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access
Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 digital
issues and online access to articles $119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be
subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR
CONTENT BEING VIEWED BY OTHERS GENOME-WIDE BIDIRECTIONAL CRISPR SCREENS IDENTIFY MUCINS AS HOST FACTORS MODULATING SARS-COV-2 INFECTION Article Open access 25 July 2022 LY6E IMPAIRS
CORONAVIRUS FUSION AND CONFERS IMMUNE CONTROL OF VIRAL DISEASE Article 23 July 2020 GENOME-WIDE CRISPR SCREENING IDENTIFIES TMEM106B AS A PROVIRAL HOST FACTOR FOR SARS-COV-2 Article 08 March
2021 DATA AVAILABILITY The authors declare that the data supporting the findings of this study are available within the article and its Supplementary Information files or are available on
request. The RNA-seq data discussed in this publication have been deposited in the Gene Expression Omnibus database (GSE209974). Source data are provided with this paper. REFERENCES *
Schoggins, J. W. Interferon-stimulated genes: what do they all do? _Annu. Rev. Virol._ 6, 567–584 (2019). CAS PubMed Google Scholar * Mao, M. et al. RIG-E, a human homolog of the murine
Ly-6 family, is induced by retinoic acid during the differentiation of acute promyelocytic leukemia cell. _Proc. Natl Acad. Sci. USA_ 93, 5910–5914 (1996). CAS PubMed PubMed Central
Google Scholar * Bacquin, A. et al. A cell fusion-based screening method identifies glycosylphosphatidylinositol-anchored protein Ly6e as the receptor for mouse endogenous retroviral
envelope syncytin-A. _J. Virol._ 91, e00832–17 (2017). CAS PubMed PubMed Central Google Scholar * Schupp, J. C. et al. Integrated single-cell atlas of endothelial cells of the human
lung. _Circulation_ 144, 286–302 (2021). CAS PubMed PubMed Central Google Scholar * Tabula Muris, C. et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris.
_Nature_ 562, 367–372 (2018). Google Scholar * Mar, K. B. et al. LY6E mediates an evolutionarily conserved enhancement of virus infection by targeting a late entry step. _Nat. Commun._ 9,
3603 (2018). PubMed PubMed Central Google Scholar * Lee, P. Y., Wang, J. X., Parisini, E., Dascher, C. C. & Nigrovic, P. A. Ly6 family proteins in neutrophil biology. _J. Leukoc.
Biol._ 94, 585–594 (2013). CAS PubMed Google Scholar * Saitoh, S. et al. Modulation of TCR-mediated signaling pathway by thymic shared antigen-1 (TSA-1)/stem cell antigen-2 (Sca-2). _J.
Immunol._ 155, 5574–5581 (1995). CAS PubMed Google Scholar * Mao, W., Hunt, H. D. & Cheng, H. H. Cloning and functional characterization of chicken stem cell antigen 2. _Dev. Comp.
Immunol._ 34, 360–368 (2010). CAS PubMed Google Scholar * Xu, X. et al. IFN-stimulated gene LY6E in monocytes regulates the CD14/TLR4 pathway but inadequately restrains the
hyperactivation of monocytes during chronic HIV-1 infection. _J. Immunol._ 193, 4125–4136 (2014). CAS PubMed Google Scholar * Classon, B. J. & Coverdale, L. Mouse stem cell antigen
Sca-2 is a member of the Ly-6 family of cell surface proteins. _Proc. Natl Acad. Sci. USA_ 91, 5296–5300 (1994). CAS PubMed PubMed Central Google Scholar * Wu, L. et al. Mouse thymus
dendritic cells: kinetics of development and changes in surface markers during maturation. _Eur. J. Immunol._ 25, 418–425 (1995). CAS PubMed Google Scholar * Noda, S., Kosugi, A., Saitoh,
S., Narumiya, S. & Hamaoka, T. Protection from anti-TCR/CD3-induced apoptosis in immature thymocytes by a signal through thymic shared antigen-1/stem cell antigen-2. _J. Exp. Med._ 183,
2355–2360 (1996). CAS PubMed Google Scholar * Zammit, D. J. et al. Essential role for the lymphostromal plasma membrane Ly-6 superfamily molecule thymic shared antigen 1 in development
of the embryonic adrenal gland. _Mol. Cell. Biol._ 22, 946–952 (2002). CAS PubMed PubMed Central Google Scholar * Langford, M. B., Outhwaite, J. E., Hughes, M., Natale, D. R. C. &
Simmons, D. G. Deletion of the Syncytin A receptor Ly6e impairs syncytiotrophoblast fusion and placental morphogenesis causing embryonic lethality in mice. _Sci. Rep._ 8, 3961 (2018). PubMed
PubMed Central Google Scholar * Krishnan, M. N. et al. RNA interference screen for human genes associated with West Nile virus infection. _Nature_ 455, 242–245 (2008). CAS PubMed
PubMed Central Google Scholar * Schoggins, J. W. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. _Nature_ 472, 481–485 (2011). CAS
PubMed PubMed Central Google Scholar * Schoggins, J. W. et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. _Nature_ 505, 691–695 (2014). CAS
PubMed Google Scholar * Hackett, B. A. & Cherry, S. Flavivirus internalization is regulated by a size-dependent endocytic pathway. _Proc. Natl Acad. Sci. USA_ 115, 4246–4251 (2018).
CAS PubMed PubMed Central Google Scholar * Yu, J., Liang, C. & Liu, S. L. Interferon-inducible LY6E protein promotes HIV-1 infection. _J. Biol. Chem._ 292, 4674–4685 (2017). CAS
PubMed PubMed Central Google Scholar * Pfaender, S. et al. LY6E impairs coronavirus fusion and confers immune control of viral disease. _Nat. Microbiol_ 5, 1330–1339 (2020). CAS PubMed
PubMed Central Google Scholar * Wickenhagen, A. et al. A prenylated dsRNA sensor protects against severe COVID-19. _Science_ 374, eabj3624 (2021). CAS PubMed PubMed Central Google
Scholar * Danziger, O., Patel, R. S., DeGrace, E. J., Rosen, M. R. & Rosenberg, B. R. Inducible CRISPR activation screen for interferon-stimulated genes identifies OAS1 as a SARS-CoV-2
restriction factor. _PLoS Pathog._ 18, e1010464 (2022). CAS PubMed PubMed Central Google Scholar * Mac Kain, A. et al. Identification of DAXX as a restriction factor of SARS-CoV-2
through a CRISPR/Cas9 screen. _Nat. Commun._ 13, 2442 (2022). Google Scholar * Abram, C. L., Roberge, G. L., Hu, Y. & Lowell, C. A. Comparative analysis of the efficiency and
specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. _J. Immunol. Methods_ 408, 89–100 (2014). CAS PubMed PubMed Central Google Scholar * McCubbrey, A. L., Allison,
K. C., Lee-Sherick, A. B., Jakubzick, C. V. & Janssen, W. J. Promoter specificity and efficacy in conditional and inducible transgenic targeting of lung macrophages. _Front. Immunol._
8, 1618 (2017). PubMed PubMed Central Google Scholar * Joseph, C. et al. Deciphering hematopoietic stem cells in their niches: a critical appraisal of genetic models, lineage tracing, and
imaging strategies. _Cell Stem Cell_ 13, 520–533 (2013). CAS PubMed Google Scholar * Bao, L. et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. _Nature_ 583, 830–833
(2020). CAS PubMed Google Scholar * Dinnon, K. H. 3rd et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. _Nature_ 586, 560–566 (2020). PubMed PubMed Central
Google Scholar * Leist, S. R. et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. _Cell_ 183, 1070–1085 e1012 (2020). CAS PubMed PubMed
Central Google Scholar * Gu, H. et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. _Science_ 369, 1603–1607 (2020). CAS PubMed PubMed Central Google Scholar *
Imai, M. et al. Characterization of a new SARS-CoV-2 variant that emerged in Brazil. _Proc. Natl Acad. Sci. USA_ 118, e2106535118 (2021). CAS PubMed PubMed Central Google Scholar *
Orthgiess, J. et al. Neurons exhibit Lyz2 promoter activity in vivo: Implications for using LysM-Cre mice in myeloid cell research. _Eur. J. Immunol._ 46, 1529–1532 (2016). CAS PubMed
Google Scholar * Gandhi, R. T., Lynch, J. B. & Del Rio, C. Mild or moderate Covid-19. _N. Engl. J. Med._ 383, 1757–1766 (2020). CAS PubMed Google Scholar * Huang, K. et al. Q493K and
Q498H substitutions in Spike promote adaptation of SARS-CoV-2 in mice. _EBioMedicine_ 67, 103381 (2021). CAS PubMed PubMed Central Google Scholar * Sun, S. et al. Characterization and
structural basis of a lethal mouse-adapted SARS-CoV-2. _Nat. Commun._ 12, 5654 (2021). CAS PubMed PubMed Central Google Scholar * Muruato, A. et al. Mouse-adapted SARS-CoV-2 protects
animals from lethal SARS-CoV challenge. _PLoS Biol._ 19, e3001284 (2021). CAS PubMed PubMed Central Google Scholar * Winkler, E. S. et al. SARS-CoV-2 infection of human ACE2-transgenic
mice causes severe lung inflammation and impaired function. _Nat. Immunol._ 21, 1327–1335 (2020). CAS PubMed PubMed Central Google Scholar * Zheng, J. et al. COVID-19 treatments and
pathogenesis including anosmia in K18-hACE2 mice. _Nature_ 589, 603–607 (2021). CAS PubMed Google Scholar * Fumagalli, V. et al. Administration of aerosolized SARS-CoV-2 to K18-hACE2 mice
uncouples respiratory infection from fatal neuroinvasion. _Sci. Immunol._ 7, eabl9929 (2022). CAS PubMed Google Scholar * Salahudeen, A. A. et al. Progenitor identification and
SARS-CoV-2 infection in human distal lung organoids. _Nature_ 588, 670–675 (2020). CAS PubMed PubMed Central Google Scholar * Ravindra, N. G. et al. Single-cell longitudinal analysis of
SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes. _PLoS Biol._ 19, e3001143 (2021). CAS PubMed PubMed Central
Google Scholar * Fiege, J. K. et al. Single cell resolution of SARS-CoV-2 tropism, antiviral responses, and susceptibility to therapies in primary human airway epithelium. _PLoS Pathog._
17, e1009292 (2021). CAS PubMed PubMed Central Google Scholar * Peng, Y. et al. Angiotensin-converting enzyme 2 in peripheral lung club cells modulates the susceptibility to SARS-CoV-2
in chronic obstructive pulmonary disease. _Am. J. Physiol. Lung Cell. Mol. Physiol._ 322, L712–L721 (2022). PubMed PubMed Central Google Scholar * Fadista, J. et al. Shared genetic
etiology between idiopathic pulmonary fibrosis and COVID-19 severity. _EBioMedicine_ 65, 103277 (2021). CAS PubMed PubMed Central Google Scholar * van Moorsel, C. H. M. et al. The MUC5B
promoter polymorphism associates with severe COVID-19 in the European population. _Front. Med._ 8, 668024 (2021). Google Scholar * Verma, A. et al. A MUC5B gene polymorphism, rs35705950-T,
confers protective effects against COVID-19 hospitalization but not severe disease or mortality. _Am. J. Respir. Crit. Care Med_ 206, 1220–1229 (2022). CAS PubMed PubMed Central Google
Scholar * Initiative, C.-H. G. A first update on mapping the human genetic architecture of COVID-19. _Nature_ 608, E1–E10 (2022). Google Scholar * Coley, S. E. et al. Recombinant mouse
hepatitis virus strain A59 from cloned, full-length cDNA replicates to high titers in vitro and is fully pathogenic in vivo. _J. Virol._ 79, 3097–3106 (2005). CAS PubMed PubMed Central
Google Scholar * Rihn, S. J. et al. A plasmid DNA-launched SARS-CoV-2 reverse genetics system and coronavirus toolkit for COVID-19 research. _PLoS Biol._ 19, e3001091 (2021). CAS PubMed
PubMed Central Google Scholar * Brattelid, T. et al. Reference gene alternatives to Gapdh in rodent and human heart failure gene expression studies. _BMC Mol. Biol._ 11, 22 (2010). PubMed
PubMed Central Google Scholar * Jhingran, A., Kasahara, S. & Hohl, T. M. Flow cytometry of lung and bronchoalveolar lavage fluid cells from mice challenged with fluorescent
_Aspergillus_ reporter (FLARE) conidia. _Bio Protoc._ 6, e1927 (2016). PubMed Google Scholar * Dietert, K. et al. Spectrum of pathogen- and model-specific histopathologies in mouse models
of acute pneumonia. _PLoS ONE_ 12, e0188251 (2017). PubMed PubMed Central Google Scholar * Choi, H. M. et al. Mapping a multiplexed zoo of mRNA expression. _Development_ 143, 3632–3637
(2016). CAS PubMed PubMed Central Google Scholar * Choi, H. M. T. et al. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust.
_Development_ 145, dev165753 (2018). PubMed PubMed Central Google Scholar * Hurskainen, M. et al. Single cell transcriptomic analysis of murine lung development on hyperoxia-induced
damage. _Nat. Commun._ 12, 1565 (2021). CAS PubMed PubMed Central Google Scholar * Richardson, R. B. et al. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that
blocks flavivirus replication. _Nat. Microbiol_ 3, 1214–1223 (2018). CAS PubMed PubMed Central Google Scholar * Shalem, O. et al. Genome-scale CRISPR–Cas9 knockout screening in human
cells. _Science_ 343, 84–87 (2014). CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We thank members of the Schoggins lab for useful discussions. We also thank M. Aufiero
and T. Hohl (Memorial Sloan Kettering Cancer Center) for advice and feedback for bone marrow chimera studies, the lab of L. Hooper (University of Texas Southwestern Medical Center (UTSW))
for CD11c-Cre and LysM-Cre transgenic mice, the labs of J. Pfeiffer (UTSW) and M. Baldridge (Washington University in St. Louis) for _Ifnar__−/−_ and _Ifnlr__−/−_ mice, respectively, the
UTSW Animal Resource Center for training and animal husbandry, the UTSW Metabolic Phenotyping Core for analysis of serum samples for ALT levels and expertise, the UTSW Histo Pathology Core,
the UTSW Preclinical Radiation Core Facility (supported by funding from CPRIT grant RP180770) and the UTSW Immunology Flow Cytometry Core. The authors also acknowledge the Quantitative Light
Microscopy Core, a Shared Resource of the Harold C. Simmons Cancer Center, supported in part by an NCI Cancer Center Support Grant, 1P30 CA142543-01, and 1S10 RR029731-01 to K. Luby-Phelps.
AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Microbiology, University of Texas Southwestern Medical Center, Dallas, TX, USA Katrina B. Mar, Alexandra I. Wells, Marley C.
Caballero Van Dyke, Alexandra H. Lopez, Jennifer L. Eitson, Wenchun Fan & John W. Schoggins * Department of Pediatrics, University of Texas Southwestern Medical Center, Dallas, TX, USA
Natasha W. Hanners * Departments of Pathology and Ophthalmology, University of Texas Southwestern Medical Center, Dallas, TX, USA Bret M. Evers * Department of Internal Medicine, University
of Texas Southwestern Medical Center, Dallas, TX, USA John M. Shelton Authors * Katrina B. Mar View author publications You can also search for this author inPubMed Google Scholar *
Alexandra I. Wells View author publications You can also search for this author inPubMed Google Scholar * Marley C. Caballero Van Dyke View author publications You can also search for this
author inPubMed Google Scholar * Alexandra H. Lopez View author publications You can also search for this author inPubMed Google Scholar * Jennifer L. Eitson View author publications You can
also search for this author inPubMed Google Scholar * Wenchun Fan View author publications You can also search for this author inPubMed Google Scholar * Natasha W. Hanners View author
publications You can also search for this author inPubMed Google Scholar * Bret M. Evers View author publications You can also search for this author inPubMed Google Scholar * John M.
Shelton View author publications You can also search for this author inPubMed Google Scholar * John W. Schoggins View author publications You can also search for this author inPubMed Google
Scholar CONTRIBUTIONS K.B.M. and J.W.S. designed the project. K.B.M., M.C.C.V.D., A.H.L. and J.W.S. performed in vivo and in vitro experiments with help from W.F. and N.W.H. A.I.W. performed
HCR-RNA-FISH and immunofluorescence experiments. J.L.E. and J.W.S. performed and analysed HBE experiments. J.L.E. performed in vitro experiments. B.M.E. and J.M.S. contributed to pathology
analysis. K.B.M. analysed remaining data and prepared figures. K.B.M. and J.W.S. wrote the manuscript. All authors reviewed and provided comments on the manuscript. This study was supported
by grants from The Clayton Foundation (to J.W.S.) and NIH (AI158124 to J.W.S. and AI132751 to N.W.H.). J.W.S. holds an Investigators in the Pathogenesis of Infectious Disease Award from the
Burroughs Wellcome Fund. CORRESPONDING AUTHOR Correspondence to John W. Schoggins. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW
INFORMATION _Nature Microbiology_ thanks Yize Li, Olivier Schwartz and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. ADDITIONAL INFORMATION
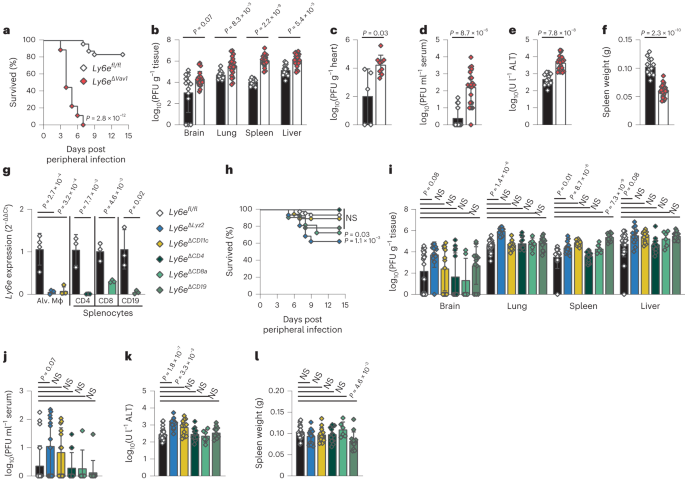
PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. EXTENDED DATA EXTENDED DATA FIG. 1 A-F, _Ly6e__fl/fl_
and _Ly6e__ΔVav1_ mice were intraperitoneally infected with 5,000 PFU MHV-A59 and assessed for survival (A), viral burden in brain, lung, spleen, and liver (B), viral burden in heart (C),
viral burden in serum (D), serum alanine aminotransferase (E), and post-mortem spleen weight (F). In A, data represents means from n = 34 _Ly6e__fl/fl_ and n = 23 Ly6eΔVav1; B, D-F, n = 8
_Ly6e__fl/fl_ and n = 15 _Ly6e__ΔVav1_; C, n = 5 _Ly6e__fl/fl_ and n = 10 _Ly6e__ΔVav1_. G, Flow cytometry gating strategy for sorting lymphocytes from the spleen for examining Ly6e gene
expression in _Ly6e__ΔCD4_, _Ly6e__ΔCD8a_, and _Ly6e__ΔCD19_ mice relative to _Ly6e__fl/fl_ littermates in Fig. 1g. H–L, mice were intraperitoneally infected with 5,000 PFU MHV-A59 and
assessed for (H) survival (n = 47 _Ly6e__fl/fl_, n = 15 _Ly6e__ΔLyz2_, n = 14 _Ly6e__ΔCD11c_, n = 14 _Ly6e__ΔCD4_, n = 5 _Ly6e__ΔCD8a_, and n = 16 _Ly6e__ΔCD19_), viral burden in brain,
lung, spleen, and liver (I), viral burden in serum (J), serum alanine aminotransferase (K), and post-mortem spleen weight (L). In I-L, data represents means from n = 30 _Ly6e__fl/fl_, n=13
_Ly6e__ΔLysM_, n = 14 _Ly6e__ΔCD11c_, n = 10 _Ly6e__ΔCD4_, n = 6 _Ly6e__ΔCD8a_, and n = 14 _Ly6e__ΔCD19_. Male and female mice were used at an approximately 1 to 1 ratio for these
experiments. Statistical significance was determined by log-rank (Mantel-Cox) tests (A, H), two-sided Mann-Whitney test (B-D), two-sided unpaired t-test (E-F), Kruskal-Wallis test (I-J), and
one-way ANOVA (K-L). Error bars represent mean ± standard deviation. EXTENDED DATA FIG. 2 A, Flow cytometry gating strategy for sorting lung CD45+ cells and CD31+ cells from _Ly6e__ΔVav1_
mice for determining Ly6e gene expression as shown in Fig. 2c,d. B, Flow cytometry gating strategy for identifying donor (CD45.1+) and recipient (CD45.2+) immune cells. C, Relative
composition of CD45.1+ and CD45.2+ immune cells of compartment total CD45+ cells in lung, spleen, and blood. D, Relative Ly6e mRNA levels in brain, heart, lung, liver, and spleen from _Ly6e_
wildtype, heterozygous, and knockout mice. In C, data represents means from n = 3 _Ly6e__fl/fl__; CD45__.2/.2_, n = 3 _Ly6e__fl/fl__; CD45__.1/.1_, n = 3 _Ly6e__fl→fl_, n = 3
_Ly6e__fl→ΔVav1_; D, n = 3 _Ly6_e wildtype, n = 3 _Ly6e_ heterozygous, n = 3 _Ly6e_ knockout. Error bars represent mean ± standard deviation. EXTENDED DATA FIG. 3 A, _Ly6e_ wildtype and
knockout mice were intranasally infected with 8,700 PFU SARS-CoV-2 and monitored daily for weight loss. Lung viral burden measured by quantitative PCR for _Ly6e_ wildtype and knockout mice
(B) or C57BL/6J, _Ifnar__−/−_, and _Ifnlr__−/−_ mice (C) that were infected with 60,000 PFU SARS-CoV-2 and euthanized 3 days post-infection. D, Lung _Ly6e_ expression from C57BL/6J,
_Ifnar__−/−_, and _Ifnlr__−/−_ mice that were mock treated with PBS or infected with 8,700 PFU SARS-CoV-2 and euthanized the next day. Expression is shown relative to PBS-treated C57BL/6J
mice. Expression of _Ly6e_ (E) and _Mx1_ (F) in lungs from C57BL/6J mice treated intranasally with recombinant human IFNλ or retro-orbitally with recombinant murine IFNβ relative to
untreated (untr). In A, data represents means from n = 5 _Ly6e_ wildtype, n = 5 _Ly6e_ knockout mice; B, n = 13 _Ly6e_ wildtype, n = 13 _Ly6e_ knockout mice; C, n = 8 C57BL6/J, n = 8
_Ifnar__−/−_, and n=8 _Ifnlr__−/−_; D, n = 5 for each group and genotype; E-F, n = 6 untreated, n = 9 IFNλ-treated, n = 8 IFNβ-treated. G, Representative hematoxylin and eosin-stained lung
sections used for analysis shown in Fig. 3e from _Ly6e_ wildtype (n = 6) and _Ly6e_ knockout (n = 6) mice euthanized 3 days after treated with PBS or intranasal infection with 60,000 PFU
SARS-CoV-2. Example of automated image analysis of SARS-CoV-2 infected cells for Fig. 3g–h (H) and of CD45+ cells for Fig. 3i, j (I). In the corresponding markup images, cells without DAB
marker (for example positive stain) are colored blue, and positive cells identified as weak positive, moderate positive, and strong positive for the DAB marker are colored as yellow, orange,
and red respectively. Statistical significance was determined by two-sided unpaired t-test (A), two-sided Mann-Whitney test (B-C), one way ANOVA with Holm-Šídák′s multiple comparisons test
(D–F). Error bars represent mean ± SEM in A and mean ± SD in B-F. Scale bars: 600 μm (G), 1 mm and 100 μm (H-I). EXTENDED DATA FIG. 4 mRNA sequencing of lungs from _Ly6e_ wildtype and _Ly6e_
knockout mice that were intranasally infected with 60,000 PFU P.1 SARS-CoV-2 and euthanized 3 days post-infection. Volcano plot summarization of differentially expressed genes (DEGs)
between uninfected _Ly6e_ wildtype (n = 6) and _Ly6e_ knockout (n = 4) mice (A), uninfected wildtype (n = 6) and SARS-CoV-2 infected wildtype (n = 7) mice (B), and uninfected knockout (n =
4) and SARS-CoV-2 infected knockout (n=7) mice (C). D, Z-score heatmap of the top 25 DEGs for data summarized in A. E, Gene Ontology (GO) enrichment analysis of transcriptomic data from
SARS-CoV-2 infected _Ly6e_ wildtype and _Ly6e_ knockout mice. The abscissa is the ratio of the number of differential genes linked with the GO term to the total number of differential genes.
The size of a point represents the number of genes annotated to a specific GO term, and the color from red to purple represents the significant level of the enrichment. Male and female mice
were used at an approximately 1 to 1 ratio for these experiments. For A-C, P-values were determined using the negative binomial distribution model and adjusted for multiple hypothesis
testing using the Benjamini and Hochberg’s approach for controlling the false discovery rate. P-values for E were determined using hypergeometric distribution and adjusted testing using the
Benjamini and Hochberg’s approach for controlling the false discovery rate. EXTENDED DATA FIG. 5 Differential expression of select genes highlighted in Fig. 4a between SARS-CoV-2-infected
_Ly6e_ wildtype mice and uninfected _Ly6e_ wildtype mice (A) and SARS-CoV-2-infected _Ly6e_ knockout mice and uninfected _Ly6e_ knockout mice (B). EXTENDED DATA FIG. 6 Mice were infected
with 8,700 PFU P.1 SARS-CoV-2 and euthanized 1 day post-infection. Lung sections were probed for SARS-CoV-2 RNA (green) and _Scgb1a1_ mRNA (red) (A, C) or _Scgb3a1_ mRNA (red) (B, D). Lung
sections from n = 3 _Ly6e_ knockout mice were stained and imaged. Images shown are from two different mice than in Fig. 4. Scale bars: 100 µM (A-B), 25 µM (C-D). EXTENDED DATA FIG. 7 Mice
were infected with 8,700 PFU SARS-CoV-2 and euthanized 1 day post-infection. Lung sections were probed for SARS-CoV-2 RNA (green) and _Sftpc_ mRNA (red) (A, C) or for SARS-CoV-2 nucleocapsid
(green) and acetylated tubulin (red) (B, D). Lung sections from n = 3 _Ly6e_ knockout mice were stained and imaged. Images shown are from different mice than in Fig. 4. Scale bars: 100 µM
(A-B), 25 µM (C-D). EXTENDED DATA FIG. 8 A, Flow cytometry gating strategy used in Fig. 4g–h for detecting SARS-CoV-2 nucleocapsid (N) in SCGB1A1-positive and acetylated tubulin-positive
pulmonary epithelial cells. Representative plots are from uninfected _Ly6e_ knockout mice. EXTENDED DATA FIG. 9 A, Area under the curve analysis of SARS-CoV-2 infectivity in HBE-ACE2 cells
from n = 3 independent experiments, B, Western blot of two independent preparations of control and LY6E knockout cells used in A. Statistical significance was determined by two-sided
unpaired t-test (A). Error bars represent mean ± standard deviation. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Tables 1–5. REPORTING SUMMARY SOURCE DATA SOURCE DATA
FIG. 1 Unprocessed western blot for Extended Dat Fig. 9. RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under
a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such
publishing agreement and applicable law. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Mar, K.B., Wells, A.I., Caballero Van Dyke, M.C. _et al._ LY6E is a pan-coronavirus
restriction factor in the respiratory tract. _Nat Microbiol_ 8, 1587–1599 (2023). https://doi.org/10.1038/s41564-023-01431-w Download citation * Received: 07 July 2022 * Accepted: 19 June
2023 * Published: 13 July 2023 * Issue Date: August 2023 * DOI: https://doi.org/10.1038/s41564-023-01431-w SHARE THIS ARTICLE Anyone you share the following link with will be able to read
this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative