Play all audios:
Efficient cytosolic delivery is a significant hurdle when using short interfering RNA (siRNA) in therapeutic applications. Here we show that cholesterol-rich exosomes are prone to entering
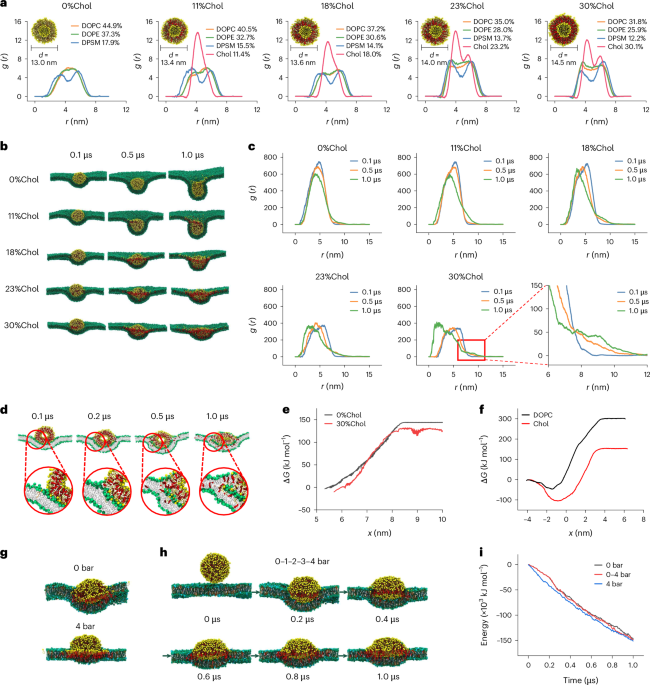
cancer cells through membrane fusion, achieving direct cytosolic delivery of siRNA. Molecular dynamics simulations suggest that deformation and increased contact with the target cell
membrane facilitate membrane fusion. In vitro we show that cholesterol-enriched milk-derived exosomes (MEs) achieve a significantly higher gene silencing effect of siRNA, inducing superior
cancer cell apoptosis compared with the native and cholesterol-depleted MEs, as well as conventional transfection agents. When administered orally or intravenously to mice bearing orthotopic
or subcutaneous tumours, the cholesterol-enriched MEs/siRNA exhibit antitumour activity superior to that of lipid nanoparticles. Collectively, by modulating the cholesterol content of
exosome membranes to facilitate cell entry via membrane fusion, we provide a promising approach for siRNA-based gene therapy, paving the way for effective, safe and simple gene therapy
strategies.
All data supporting the findings of this study are included in the article and the Supplementary Information. There are no data from third-party or publicly available datasets. All data
generated as part of this study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
We sincerely appreciate the financial support from the National Science Fund of Distinguished Young Scholars (82025032), the National Key Research and Development Program of China
(2022YFA1203200), the National Natural Science Foundation of China (82073773), the Young Elite Scientists Sponsorship Program by CAST (2022QNRC001), the Key Research Program of Chinese
Academy of Sciences (ZDBS-ZRKJZ-TLC005), the ‘Open Competition to Select the Best Candidates’ Key Technology Program for Nucleic Acid Drugs of NCTIB (NCTIB2022HS01006), the Shanghai Action
Plan for Science, Technology and Innovation (23HC1401200) and the Shanghai Institute of Materia Medica, Chinese Academy of Sciences (SIMM0220232001). We thank the staff members of the
Integrated Laser Microscopy System, the Electron Microscopy System, the Molecular Imaging System and the Large-scale Protein Preparation System at the National Facility for Protein Science
in Shanghai (NFPS), Shanghai Advanced Research Institute, Chinese Academy of Sciences, China, for sample preparation, data collection and analysis. The MD simulations were performed on
BSCC-A6 at the Beijing Super Cloud Computing Center. We are grateful to G. Li, Y. Yu, F. Gong and F. Liu for cryo-TEM data collection, cell imaging and AFM data collection, respectively.
State Key Laboratory of Drug Research and Center of Pharmaceutics, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China
Yan Zhuo, Zhu Zhu, Jie Wang, Xiang Li, Zhuan Zhang, Cong Guo, Bingqi Wang, Di Nie, Yong Gan & Miaorong Yu
School of Pharmacy, Jiangxi Medical College, Nanchang University, Nanchang, China
Department of Engineering Mechanics, State Key Laboratory of Fluid Power and Mechatronic Systems, Zhejiang University, Hangzhou, China
School of Pharmacy, Shanghai University of Traditional Chinese Medicine, Shanghai, China
Xiang Li, Cong Guo, Bingqi Wang, Di Nie, Yong Gan & Miaorong Yu
NMPA Key Laboratory or Quality Research and Evaluation of Pharmaceutical Excipients, National Institutes for Food and Drug Control, Beijing, China
M.Y., G.H., Y.G., Y.Z. and Z.L. planned and executed the experiments, analysed the data and were involved in discussions of the data. M.Y., G.H., Y.Z., Z.L. and Z. Zhu wrote the paper. Y.Z.,
Z.L., Z. Zhu, J.W., X.L., Z. Zhang, C.G., B.W. and D.N. performed the experiments. All authors critically reviewed and approved the paper.
Nature Nanotechnology thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Discussion, Methods, Table 1, Figs. 1–22 and References.
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author
self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Anyone you share the following link with will be able to read this content: