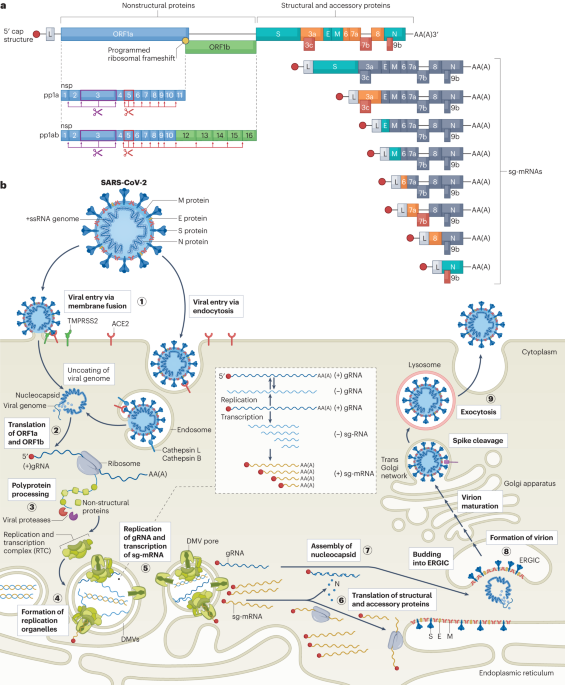
Play all audios:
ABSTRACT The zoonotic emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the ensuing coronavirus disease 2019 (COVID-19) pandemic have profoundly affected our
society. The rapid spread and continuous evolution of new SARS-CoV-2 variants continue to threaten global public health. Recent scientific advances have dissected many of the molecular and
cellular mechanisms involved in coronavirus infections, and large-scale screens have uncovered novel host-cell factors that are vitally important for the virus life cycle. In this Review, we
provide an updated summary of the SARS-CoV-2 life cycle, gene function and virus–host interactions, including recent landmark findings on general aspects of coronavirus biology and newly
discovered host factors necessary for virus replication. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS
OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn
more Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to
full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our
FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS CELLULAR HOST FACTORS FOR SARS-COV-2 INFECTION Article 01 September 2021 CORONAVIRUS BIOLOGY AND REPLICATION:
IMPLICATIONS FOR SARS-COV-2 Article 28 October 2020 GENOME-WIDE BIDIRECTIONAL CRISPR SCREENS IDENTIFY MUCINS AS HOST FACTORS MODULATING SARS-COV-2 INFECTION Article Open access 25 July 2022
REFERENCES * Gorbalenya, A. E. et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. _Nat. Microbiol._ 5, 536–544 (2020).
Google Scholar * Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. _Nature_ 579, 270–273 (2020). CAS PubMed ADS PubMed Central Google
Scholar * Zhu, N. et al. A novel coronavirus from patients with pneumonia in China, 2019. _N. Engl. J. Med._ 382, 727–733 (2020). CAS PubMed PubMed Central Google Scholar * Coronavirus
(COVID-19) Dashboard. _WHO_ https://covid19.who.int/ (2022). * Telenti, A., Hodcroft, E. B. & Robertson, D. L. The evolution and biology of SARS-CoV-2 variants. _Cold Spring Harb. Persp.
Med._ 12, a041390 (2022). CAS Google Scholar * Harvey, W. T. et al. SARS-CoV-2 variants, spike mutations and immune escape. _Nat. Rev. Microbiol._ 19, 409–424 (2021). CAS PubMed PubMed
Central Google Scholar * Grant, R. et al. When to update COVID-19 vaccine composition. _Nat. Med._ 29, 776–780 (2023). CAS PubMed Google Scholar * Jungreis, I., Sealfon, R. &
Kellis, M. SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 _Sarbecovirus_ genomes. _Nat. Commun._ 12, 2642 (2021). CAS PubMed ADS PubMed Central Google Scholar *
Jungreis, I. et al. Conflicting and ambiguous names of overlapping ORFs in the SARS-CoV-2 genome: a homology-based resolution. _Virology_ 558, 145–151 (2021). CAS PubMed Google Scholar *
Finkel, Y. et al. The coding capacity of SARS-CoV-2. _Nature_ 589, 125–130 (2020). PubMed ADS Google Scholar * Gordon, D. E. et al. A SARS-CoV-2 protein interaction map reveals targets
for drug repurposing. _Nature_ 583, 459–468 (2020). CAS PubMed ADS PubMed Central Google Scholar * Kim, D. et al. The architecture of SARS-CoV-2 transcriptome. _Cell_ 181, 914–921.e10
(2020). CAS PubMed PubMed Central Google Scholar * Huston, N. C. et al. Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms.
_Mol. Cell_ 81, 584–598.e5 (2021). CAS PubMed PubMed Central Google Scholar * Lan, T. C. T. et al. Secondary structural ensembles of the SARS-CoV-2 RNA genome in infected cells. _Nat.
Commun._ 13, 1128 (2022). CAS PubMed ADS PubMed Central Google Scholar * Ziv, O. et al. The short- and long-range RNA–RNA interactome of SARS-CoV-2. _Mol. Cell_ 80, 1067–1077.e5 (2020).
CAS PubMed PubMed Central Google Scholar * Madhugiri, R., Fricke, M., Marz, M. & Ziebuhr, J. Coronavirus _cis_-acting RNA elements. _Adv. Virus Res._ 96, 127–163 (2016). CAS PubMed
PubMed Central Google Scholar * Tidu, A. et al. The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. _RNA_ 27,
253–264 (2021). THIS PUBLICATION DEMONSTRATED THAT SARS-COV-2 RELIES ON STEM LOOP 1 IN THE 5′ UTR TO EVADE THE NSP1-INDUCED TRANSLATIONAL SHUTOFF OF ITS OWN GENES. CAS PubMed Central
Google Scholar * Bujanic, L. et al. The key features of SARS-CoV-2 leader and NSP1 required for viral escape of NSP1-mediated repression. _RNA_ 28, 766–779 (2022). CAS PubMed PubMed
Central Google Scholar * Iserman, C. et al. Genomic RNA elements drive phase separation of the SARS-CoV-2 nucleocapsid. _Mol. Cell_ 80, 1078–1091.e6 (2020). CAS PubMed PubMed Central
Google Scholar * Bhatt, P. R. et al. Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. _Science_ 372, 1306–1313 (2021). IN-DEPTH STRUCTURAL AND
BIOCHEMICAL ANALYSIS INTO THE MECHANISM OF THE PROGRAMMED RIBOSOMAL FRAMESHIFT FOR SARS-COV-2. CAS PubMed ADS PubMed Central Google Scholar * Sun, L. et al. In vivo structural
characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs. _Cell_ 184, 1865–1883.e20 (2021). CAS PubMed PubMed Central Google Scholar *
Jackson, C. B., Farzan, M., Chen, B. & Choe, H. Mechanisms of SARS-CoV-2 entry into cells. _Nat. Rev. Mol. Cell Biol._ 23, 3–20 (2021). COMPREHENSIVE REVIEW ON SARS-COV-2 ENTRY
MECHANISM. PubMed PubMed Central Google Scholar * Hoffmann, M., Kleine-Weber, H. & Pöhlmann, S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for
infection of human lung cells. _Mol. Cell_ 78, 779–784.e5 (2020). THIS ARTICLE HIGHLIGHTS THE PRESENCE OF A MULTIBASIC S1/S2 CLEAVAGE SITE IN THE SARS-COV-2 SPIKE PROTEIN THAT CAN BE CUT BY
FURIN AND IS A PREREQUISITE FOR VIRAL ENTRY INTO LUNG CELLS. CAS PubMed PubMed Central Google Scholar * Hansen, J. et al. Studies in humanized mice and convalescent humans yield a
SARS-CoV-2 antibody cocktail. _Science_ 369, 1010–1014 (2020). CAS PubMed ADS PubMed Central Google Scholar * Robbiani, D. F. et al. Convergent antibody responses to SARS-CoV-2 in
convalescent individuals. _Nature_ 584, 437–442 (2020). CAS PubMed ADS PubMed Central Google Scholar * Pinto, D. et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV
antibody. _Nature_ 583, 290–295 (2020). CAS PubMed ADS Google Scholar * Yuan, M. et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV.
_Science_ 368, 630–633 (2020). CAS PubMed ADS PubMed Central Google Scholar * Liu, L. et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. _Nature_ 584,
450–456 (2020). THIS IS ONE OF THE FIRST PUBLICATIONS TO REPORT THE RECEPTOR-BINDING DOMAIN (RBD) AND N-TERMINAL DOMAIN (NTD) EPITOPES AS THE TWO MAIN NEUTRALIZATION TARGETS ON THE
SARS-COV-2 SPIKE PROTEIN. CAS PubMed Google Scholar * Chi, X. et al. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. _Science_ 369,
650–655 (2020). CAS PubMed ADS PubMed Central Google Scholar * Meng, B. et al. SARS-CoV-2 spike N-terminal domain modulates TMPRSS2-dependent viral entry and fusogenicity. _Cell Rep._
40, 111220 (2022). HERE, IT WAS SHOWN THAT THE SARS-COV-2 SPIKE PROTEIN’S NTD CAN MODULATE S1/S2 CLEAVAGE AND INFLUENCE TMPRSS2 USAGE AND FUSOGENICITY. CAS PubMed PubMed Central Google
Scholar * Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. _Cell_ 181, 271–280.e8 (2020). THE FIRST
PUBLICATION TO CONFIRM THAT, SIMILAR TO SARS-COV, THE PROCESSING OF THE SARS-COV-2 SPIKE PROTEIN IS MEDIATED BY TMPRSS2. CAS PubMed PubMed Central Google Scholar * Zhao, M. M. et al.
Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. _Signal. Transduct. Target. Ther._ 6, 134 (2021). CAS
PubMed PubMed Central Google Scholar * Ziegler, C. G. K. et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell
subsets across tissues. _Cell_ 181, 1016–1035.e19 (2020). CAS PubMed PubMed Central Google Scholar * Su, M. C. et al. An atypical RNA pseudoknot stimulator and an upstream attenuation
signal for −1 ribosomal frameshifting of SARS coronavirus. _Nucleic Acids Res._ 33, 4265–4275 (2005). CAS PubMed PubMed Central Google Scholar * Zhang, K. et al. Cryo-EM and antisense
targeting of the 28-kDa frameshift stimulation element from the SARS-CoV-2 RNA genome. _Nat. Struct. Mol. Biol._ 28, 747–754 (2021). CAS PubMed PubMed Central Google Scholar * Brierley,
I., Digard, P. & Inglis, S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. _Cell_ 57, 537–547 (1989). CAS PubMed
PubMed Central Google Scholar * Sun, Y. et al. Restriction of SARS-CoV-2 replication by targeting programmed –1 ribosomal frameshifting. _Proc. Natl Acad. Sci. USA_ 118, e2023051118
(2021). CAS PubMed PubMed Central Google Scholar * Osipiuk, J. et al. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. _Nat. Commun._ 12,
743 (2021). CAS PubMed ADS PubMed Central Google Scholar * Jin, Z. et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. _Nature_ 582, 289–293 (2020). CAS PubMed
ADS Google Scholar * Ziebuhr, J., Snijder, E. J. & Gorbalenya, A. E. Virus-encoded proteinases and proteolytic processing in the _Nidovirales_. _J. Gen. Virol._ 81, 853–879 (2000). CAS
PubMed Google Scholar * Thoms, M. et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. _Science_ 369, 1249–1256 (2020). THOMS ET AL.
(2020) AND SCHUBERT ET AL. (2020) ELUCIDATE THE BINDING OF SARS-COV-2 NSP1 TO THE RIBOSOME AND CAUSE TRANSLATIONAL SHUTDOWN. CAS PubMed ADS PubMed Central Google Scholar * Schubert, K.
et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. _Nat. Struct. Mol. Biol._ 27, 959–966 (2020). CAS PubMed Google Scholar * Fisher, T. et al. Parsing the
role of NSP1 in SARS-CoV-2 infection. _Cell Rep._ 39, 110954 (2022). CAS PubMed PubMed Central Google Scholar * Snijder, E. J., Decroly, E. & Ziebuhr, J. The nonstructural proteins
directing coronavirus RNA synthesis and processing. In _Advances in Virus Research_ Vol. 96 (ed. Ziebuhr, J.) 59–126 (Academic Press, 2016). * Cortese, M. et al. Integrative imaging reveals
SARS-CoV-2-induced reshaping of subcellular morphologies. _Cell Host Microbe_ 28, 853–866.e5 (2020). CAS PubMed PubMed Central Google Scholar * Snijder, E. J. et al. A unifying
structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. _PLoS Biol._ 18, e3000715 (2020). CAS PubMed PubMed Central Google Scholar * Wolff,
G., Melia, C. E., Snijder, E. J. & Bárcena, M. Double-membrane vesicles as platforms for viral replication. _Trends Microbiol._ 28, 1022–1033 (2020). CAS PubMed PubMed Central Google
Scholar * Klein, S. et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. _Nat. Commun._ 11, 5885 (2020). CAS PubMed ADS PubMed Central Google
Scholar * Ricciardi, S. et al. The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. _Nature_ 606, 761–768 (2022). CAS PubMed ADS PubMed Central Google Scholar *
Twu, W. I. et al. Contribution of autophagy machinery factors to HCV and SARS-CoV-2 replication organelle formation. _Cell Rep._ 37, 110049 (2021). CAS PubMed PubMed Central Google
Scholar * Tabata, K. et al. Convergent use of phosphatidic acid for hepatitis C virus and SARS-CoV-2 replication organelle formation. _Nat. Commun._ 12, 7276 (2021). CAS PubMed ADS
PubMed Central Google Scholar * Ji, M. et al. VMP1 and TMEM41B are essential for DMV formation during β-coronavirus infection. _J. Cell Biol._ 221, e202112081 (2022). PubMed PubMed
Central Google Scholar * Wolff, G. et al. A molecular pore spans the double membrane of the coronavirus replication organelle. _Science_ 369, 1395–1398 (2020). CAS PubMed ADS PubMed
Central Google Scholar * Zimmermann, L. et al. SARS-CoV-2 nsp3 and nsp4 are minimal constituents of a pore spanning replication organelle. _Nat. Commun._ 14, 7894 (2023). CAS PubMed ADS
PubMed Central Google Scholar * da Silva Gomes Dias, S. et al. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. _PLoS Pathog._ 16, e1009127 (2020).
Google Scholar * Malone, B., Urakova, N., Snijder, E. J. & Campbell, E. A. Structures and functions of coronavirus replication–transcription complexes and their relevance for SARS-CoV-2
drug design. _Nat. Rev. Mol. Cell Biol._ 23, 21–39 (2021). PubMed PubMed Central Google Scholar * Gao, Y. et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
_Science_ 368, 779–782 (2020). CAS PubMed ADS PubMed Central Google Scholar * Wang, Q. et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. _Cell_ 23, 182–417.e13
(2020). Google Scholar * Hillen, H. S. et al. Structure of replicating SARS-CoV-2 polymerase. _Nature_ 584, 154–156 (2020). CAS PubMed ADS Google Scholar * Mickolajczyk, K. J. et al.
Force-dependent stimulation of RNA unwinding by SARS-CoV-2 nsp13 helicase. _Biophys. J._ 120, 1020–1030 (2021). CAS PubMed ADS Google Scholar * Chen, J. et al. Structural basis for
helicase-polymerase coupling in the SARS-CoV-2 replication–transcription complex. _Cell_ 182, 1560–1573.e13 (2020). CAS PubMed PubMed Central Google Scholar * Yan, L. et al. Architecture
of a SARS-CoV-2 mini replication and transcription complex. _Nat. Commun._ 11, 5874 (2020). CAS PubMed ADS PubMed Central Google Scholar * Malone, B. et al. Structural basis for
backtracking by the SARS-CoV-2 replication–transcription complex. _Proc. Natl Acad. Sci. USA_ 118, e2102516118 (2021). CAS PubMed PubMed Central Google Scholar * Chen, J. et al. Ensemble
cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication–transcription complex. _Nat. Struct. Mol. Biol._ 29, 250–260 (2022). MathSciNet CAS
PubMed PubMed Central Google Scholar * Nudler, E. RNA polymerase backtracking in gene regulation and genome instability. _Cell_ 149, 1438–1445 (2012). CAS PubMed Google Scholar *
Shannon, A. et al. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. _Nat. Commun._ 11, 4682 (2020). CAS
PubMed ADS PubMed Central Google Scholar * Smith, E. C., Blanc, H., Vignuzzi, M. & Denison, M. R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal
mutagenesis: evidence for proofreading and potential therapeutics. _PLoS Pathog._ 9, e1003565 (2013). CAS PubMed PubMed Central Google Scholar * Lin, S. et al. Crystal structure of
SARS-CoV-2 nsp10 bound to nsp14–ExoN domain reveals an exoribonuclease with both structural and functional integrity. _Nucleic Acids Res._ 49, 5382–5392 (2021). CAS PubMed ADS PubMed
Central Google Scholar * Liu, C. et al. Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme. _Science_ 373, 1142–1146 (2021). CAS PubMed ADS PubMed Central
Google Scholar * Yan, L. et al. Coupling of N7-methyltransferase and 3′-5′ exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading. _Cell_ 184,
3474–3485.e11 (2021). CAS PubMed PubMed Central Google Scholar * Sawicki, S. G. & Sawicki, D. L. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative
strands. In _Advances in Experimental Medicine and Biology_ Vol. 380 (eds Talbot, P. J. & Levy, G. A.) 499–506 (Springer, 1995). * Wang, D. et al. The SARS-CoV-2 subgenome landscape and
its novel regulatory features. _Mol. Cell_ 81, 2135–2147.e5 (2021). CAS PubMed PubMed Central Google Scholar * Sola, I., Almazán, F., Zúñiga, S. & Enjuanes, L. Continuous and
discontinuous RNA synthesis in coronaviruses. _Annu. Rev. Virol._ 2, 265–288 (2015). CAS PubMed PubMed Central Google Scholar * Zhang, Y. et al. In vivo structure and dynamics of the
SARS-CoV-2 RNA genome. _Nat. Commun._ 12, 5695 (2021). CAS PubMed ADS PubMed Central Google Scholar * Mendez, A. S. et al. The N-terminal domain of SARS-CoV-2 nsp1 plays key roles in
suppression of cellular gene expression and preservation of viral gene expression. _Cell Rep._ 37, 109841 (2021). CAS PubMed PubMed Central Google Scholar * Vora, S. M. et al. Targeting
stem-loop 1 of the SARS-CoV-2 50 UTR to suppress viral translation and Nsp1 evasion. _Proc. Natl Acad. Sci. USA_ 119, e2117198119 (2022). CAS PubMed PubMed Central Google Scholar *
Finkel, Y. et al. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. _Nature_ 594, 240–245 (2021). CAS PubMed ADS Google Scholar * Walker, A. P. et al. The
SARS-CoV-2 RNA polymerase is a viral RNA capping enzyme. _Nucleic Acids Res._ 49, 13019–13030 (2021). THESE AUTHORS IDENTIFY THE NIDOVIRUS RDRP-ASSOCIATED NUCLEOTIDYLTRANSFERASE (NIRAN)
DOMAIN OF NSP12 AS A CAPPING ENZYME INVOLVED IN THE FORMATION OF THE CAP CORE STRUCTURE. CAS PubMed PubMed Central Google Scholar * Pan, R. et al. N7-methylation of the coronavirus RNA
cap is required for maximal virulence by preventing innate immune recognition. _mBio_ 13, e0366221 (2022). PubMed Google Scholar * Russ, A. et al. Nsp16 shields SARS–CoV-2 from efficient
MDA5 sensing and IFIT1-mediated restriction. _EMBO Rep._ 23, e55648 (2022). CAS PubMed PubMed Central Google Scholar * Decroly, E., Ferron, F., Lescar, J. & Canard, B. Conventional
and unconventional mechanisms for capping viral mRNA. _Nat. Rev. Microbiol._ 10, 51–65 (2012). CAS Google Scholar * Kikkert, M. Innate immune evasion by human respiratory RNA viruses. _J.
Innate Immun._ 12, 4–20 (2020). CAS PubMed Google Scholar * Chen, Y. et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase.
_Proc. Natl Acad. Sci._ 106, 3484–3489 (2009). CAS PubMed ADS PubMed Central Google Scholar * Bouvet, M. et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. _PLoS
Pathog._ 6, e1000863 (2010). PubMed PubMed Central Google Scholar * Lehmann, K. C. et al. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved
domain in the RNA polymerase-containing protein of all nidoviruses. _Nucleic Acids Res._ 43, 8416–8434 (2015). CAS PubMed PubMed Central Google Scholar * Slanina, H. et al. Coronavirus
replication-transcription complex: vital and selective NMPylation of a conserved site in nsp9 by the NiRAN–RdRp subunit. _Proc. Natl Acad. Sci. USA_ 118, e2022310118 (2021). CAS PubMed
PubMed Central Google Scholar * Yan, L. et al. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. _Cell_ 184,
184–193.e10 (2021). CAS PubMed Google Scholar * Park, G. J. et al. The mechanism of RNA capping by SARS-CoV-2. _Nature_ 609, 793–800 (2022). THESE AUTHORS SHOW THE RNAYLATION OF NSP9 BY
NIRAN IN VITRO AND HENCE PROPOSE AN UNCONVENTIONAL CAPPING MECHANISM FOR SARS-COV-2. CAS PubMed ADS PubMed Central Google Scholar * Yan, L. et al. A mechanism for SARS-CoV-2 RNA capping
and its inhibition by nucleotide analog inhibitors. _Cell_ 185, 4347–4360.e17 (2022). CAS PubMed PubMed Central Google Scholar * Ogino, T. & Green, T. J. RNA synthesis and capping
by non-segmented negative strand RNA viral polymerases: lessons from a prototypic virus. _Front. Microbiol._ 10, 1490 (2019). PubMed PubMed Central Google Scholar * Wang, B., Svetlov, D.
& Artsimovitch, I. NMPylation and de-NMPylation of SARS-CoV-2 nsp9 by the NiRAN domain. _Nucleic Acids Res._ 49, 8822–8835 (2021). CAS PubMed PubMed Central Google Scholar * Cong, Y.
et al. Nucleocapsid protein recruitment to replication–transcription complexes plays a crucial role in coronaviral life cycle. _J. Virol._ 94, e01925–e02019 (2020). CAS PubMed PubMed
Central Google Scholar * Cubuk, J. et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. _Nat. Commun._ 12, 1936 (2021). CAS PubMed ADS PubMed
Central Google Scholar * Lu, S. et al. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. _Nat. Commun._ 12,
502 (2021). CAS PubMed ADS PubMed Central Google Scholar * Yao, H. et al. Molecular architecture of the SARS-CoV-2 virus. _Cell_ 183, 730–738.e13 (2020). CAS PubMed PubMed Central
Google Scholar * Bracquemond, D. & Muriaux, D. Betacoronavirus assembly: clues and perspectives for elucidating SARS-CoV-2 particle formation and egress. _mBio_ 12, e0237121 (2021).
PubMed Google Scholar * Cascarina, S. M. & Ross, E. D. Phase separation by the SARS-CoV-2 nucleocapsid protein: consensus and open questions. _J. Biol. Chem._ 298, 101677 (2022). CAS
PubMed PubMed Central Google Scholar * Boson, B. et al. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of
virus-like particles. _J. Biol. Chem._ 296, 100111 (2021). CAS PubMed Google Scholar * de Haan, C. A. M. & Rottier, P. J. M. Molecular interactions in the assembly of coronaviruses.
_Adv. Virus Res._ 64, 165–230 (2005). PubMed PubMed Central Google Scholar * Mandala, V. S. et al. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in
lipid bilayers. _Nat. Struct. Mol. Biol._ 27, 1202–1208 (2020). CAS PubMed PubMed Central Google Scholar * Nieto-Torres, J. L. et al. Severe acute respiratory syndrome coronavirus E
protein transports calcium ions and activates the NLRP3 inflammasome. _Virology_ 485, 330–339 (2015). CAS PubMed Google Scholar * Ghosh, S. et al. β-Coronaviruses use lysosomes for egress
instead of the biosynthetic secretory pathway. _Cell_ 183, 1520–1535.e14 (2020). THIS PAPER FIRST PROPOSED AND INVESTIGATED THE EGRESS OF SARS-COV-2 AND OTHER BETACORONAVIRUSES VIA
LYSOSOMAL TRAFFICKING. CAS PubMed PubMed Central Google Scholar * Tancini, B. et al. Lysosomal exocytosis: the extracellular role of an intracellular organelle. _Membranes_ 10, 406
(2020). CAS PubMed PubMed Central Google Scholar * Pu, J. et al. BORC, a multisubunit complex that regulates lysosome positioning. _Dev. Cell_ 33, 176–188 (2015). CAS PubMed PubMed
Central Google Scholar * Chen, D. et al. ORF3a of SARS-CoV-2 promotes lysosomal exocytosis-mediated viral egress. _Dev. Cell_ 56, 3250–3263.e5 (2021). CAS PubMed PubMed Central Google
Scholar * Minkoff, J. M. & tenOever, B. Innate immune evasion strategies of SARS-CoV-2. _Nat. Rev. Microbiol._ 21, 178–194 (2023). COMPREHENSIVE REVIEW ON SARS-COV-2 INNATE IMMUNE
EVASION. CAS PubMed PubMed Central Google Scholar * Gao, B. et al. Inhibition of anti-viral stress granule formation by coronavirus endoribonuclease nsp15 ensures efficient virus
replication. _PLoS Pathog._ 17, e1008690 (2021). CAS PubMed PubMed Central Google Scholar * Banerjee, A. K. et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to
suppress host defenses. _Cell_ 183, 1325–1339.e21 (2020). THEY IDENTIFIED THE VIRAL PROTEINS INVOLVED (NSP1, NAP8, NSP9 AND NSP16) IN GLOBAL INHIBITION OF HOST MRNA SPLICING, PROTEIN
TRANSLATION AND MEMBRANE PROTEIN TRAFFICKING. CAS PubMed PubMed Central Google Scholar * Hayn, M. et al. Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate
immune antagonists and remaining vulnerabilities. _Cell Rep._ 35, 109126 (2021). CAS PubMed PubMed Central Google Scholar * Lei, X. et al. Activation and evasion of type I interferon
responses by SARS-CoV-2. _Nat. Commun._ 11, 3812 (2020). ADS Google Scholar * Li, J. Y. et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling
pathway. _Virus Res._ 286, 198074 (2020). CAS PubMed Google Scholar * Shemesh, M. et al. SARS-CoV-2 suppresses IFNβ production mediated by NSP1, 5, 6, 15, ORF6 and ORF7b but does not
suppress the effects of added interferon. _PLoS Pathog._ 17, e1009800 (2021). CAS PubMed PubMed Central Google Scholar * Stukalov, A. et al. Multilevel proteomics reveals host
perturbations by SARS-CoV-2 and SARS-CoV. _Nature_ 594, 246–252 (2021). CAS PubMed ADS Google Scholar * Vazquez, C. et al. SARS-CoV-2 viral proteins NSP1 and NSP13 inhibit interferon
activation through distinct mechanisms. _PLoS One_ 16, e0253089 (2021). CAS PubMed PubMed Central Google Scholar * Xia, H. et al. Evasion of type I interferon by SARS-CoV-2. _Cell Rep._
33, 108234 (2020). ONE OF THE FIRST PAPERS TO REPORT THE INTERFERON ANTAGONISM OF SARS-COV-2. CAS PubMed PubMed Central Google Scholar * Yuen, C. K. et al. SARS-CoV-2 nsp13, nsp14, nsp15
and orf6 function as potent interferon antagonists. _Emerg. Microbes Infect._ 9, 1418–1428 (2020). CAS PubMed PubMed Central Google Scholar * Zheng, M. et al. TLR2 senses the SARS-CoV-2
envelope protein to produce inflammatory cytokines. _Nat. Immunol._ 22, 829–838 (2021). CAS PubMed PubMed Central Google Scholar * Planès, R., Bert, J. B., Tairi, S., Benmohamed, L.
& Bahraoui, E. SARS-CoV-2 envelope (E) protein binds and activates TLR2 pathway: a novel molecular target for COVID-19 interventions. _Viruses_ 14, 999 (2022). PubMed PubMed Central
Google Scholar * Moreno-Eutimio, M. A., López-Macías, C. & Pastelin-Palacios, R. Bioinformatic analysis and identification of single-stranded RNA sequences recognized by TLR7/8 in the
SARS-CoV-2, SARS-CoV, and MERS-CoV genomes. _Microbes Infect._ 22, 226–229 (2020). CAS PubMed PubMed Central Google Scholar * Choudhury, A. & Mukherjee, S. In silico studies on the
comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. _J. Med. Virol._ 92, 2105–2113 (2020). CAS PubMed PubMed
Central Google Scholar * Bortolotti, D. et al. Tlr3 and tlr7 RNA sensor activation during SARS-CoV-2 infection. _Microorganisms_ 9, 1820 (2021). CAS PubMed PubMed Central Google Scholar
* Rebendenne, A. et al. SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. _J. Virol._ 95, e02415–e02420 (2021).
CAS PubMed PubMed Central Google Scholar * Thorne, L. G. et al. SARS‐CoV‐2 sensing by RIG‐I and MDA5 links epithelial infection to macrophage inflammation. _EMBO J._ 40, e107826 (2021).
CAS PubMed PubMed Central Google Scholar * Yang, D.-M., Geng, T.-T., Harrison, A. G. & Wang, P.-H. Differential roles of RIG-I like receptors in SARS-CoV-2 infection. _Milit. Med.
Res._ 8, 49 (2021). CAS Google Scholar * Yin, X. et al. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. _Cell Rep._ 34, 108628 (2021). CAS PubMed PubMed
Central Google Scholar * Frazier, M. N. et al. Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease. _Nucleic Acids Res._ 50, 8290–8301 (2022). CAS
PubMed PubMed Central Google Scholar * Hackbart, M., Deng, X. & Baker, S. C. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. _Proc.
Natl Acad. Sci. USA_ 117, 8094–8103 (2020). CAS PubMed ADS PubMed Central Google Scholar * Ancar, R. et al. Physiologic RNA targets and refined sequence specificity of coronavirus
EndoU. _RNA_ 26, 1976–1999 (2020). CAS PubMed PubMed Central Google Scholar * Singh, K. K., Chaubey, G., Chen, J. Y. & Suravajhala, P. Decoding SARS-CoV-2 hijacking of host
mitochondria in COVID-19 pathogenesis. _Am. J. Physiol. Cell Physiol._ 319, C258–C267 (2020). PubMed PubMed Central Google Scholar * Schoggins, J. W. et al. Pan-viral specificity of
IFN-induced genes reveals new roles for cGAS in innate immunity. _Nature_ 505, 691–695 (2014). CAS PubMed ADS Google Scholar * Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic
GMP–AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. _Science_ 339, 786–791 (2013). CAS PubMed ADS Google Scholar * Rui, Y. et al. Unique and
complementary suppression of cGAS-STING and RNA sensing — triggered innate immune responses by SARS-CoV-2 proteins. _Signal. Transduct. Target. Ther._ 6, 123 (2021). CAS PubMed PubMed
Central Google Scholar * Humphries, F. et al. A diamidobenzimidazole STING agonist protects against SARS-CoV-2 infection. _Sci. Immunol._ 6, eabi9002 (2021). PubMed PubMed Central Google
Scholar * Li, M. et al. Pharmacological activation of STING blocks SARS-CoV-2 infection. _Sci. Immunol._ 6, eabi9007 (2021). PubMed PubMed Central Google Scholar * Christgen, S. &
Kanneganti, T.-D. Inflammasomes and the fine line between defense and disease. _Curr. Opin. Immunol._ 62, 39–44 (2020). CAS PubMed Google Scholar * Kelley, N., Jeltema, D., Duan, Y. &
He, Y. The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. _Int. J. Mol. Sci. 2019_ 20, 3328 (2019). CAS Google Scholar * Campbell, G. R., To, R. K., Hanna, J.
& Spector, S. A. SARS-CoV-2, SARS-CoV-1, and HIV-1 derived ssRNA sequences activate the NLRP3 inflammasome in human macrophages through a non-classical pathway. _iScience_ 24, 102295
(2021). CAS PubMed ADS PubMed Central Google Scholar * Pan, P. et al. SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. _Nat. Commun._ 12, 4664
(2021). CAS PubMed ADS PubMed Central Google Scholar * Xu, H. et al. SARS-CoV-2 viroporin encoded by ORF3a triggers the NLRP3 inflammatory pathway. _Virology_ 568, 13–22 (2022). CAS
PubMed Google Scholar * Rodrigues, T. S. et al. Inflammasomes are activated in response to SARS-cov-2 infection and are associated with COVID-19 severity in patients. _J. Exp. Med_. 218,
e20201707 (2020). PubMed Central Google Scholar * Yalcinkaya, M. et al. Modulation of the NLRP3 inflammasome by Sars-CoV-2 Envelope protein. _Sci. Rep._ 11, 24432 (2021). CAS PubMed ADS
PubMed Central Google Scholar * Zhang, Y. et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. _Proc. Natl Acad. Sci. USA_ 118, e2024202118
(2021). CAS PubMed PubMed Central Google Scholar * Arshad, N. et al. SARS-CoV-2 accessory proteins ORF7a and ORF3a use distinct mechanisms to down-regulate MHC-I surface expression.
_Proc. Natl Acad. Sci. USA_ 120, e2208525120 (2023). CAS PubMed Google Scholar * Yoo, J. S. et al. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the
STAT1-IRF1-NLRC5 axis. _Nat. Commun._ 12, 6602 (2021). CAS PubMed ADS PubMed Central Google Scholar * Menachery, V. D. et al. MERS-CoV and H5N1 influenza virus antagonize antigen
presentation by altering the epigenetic landscape. _Proc. Natl Acad. Sci. USA_ 115, E1012–E1021 (2018). CAS PubMed PubMed Central Google Scholar * Kee, J. et al. SARS-CoV-2 disrupts host
epigenetic regulation via histone mimicry. _Nature_ 610, 381–388 (2022). CAS PubMed ADS PubMed Central Google Scholar * Kim, Y. M. & Shin, E. C. Type I and III interferon responses
in SARS-CoV-2 infection. _Exp. Mol. Med._ 53, 750–760 (2021). CAS PubMed PubMed Central Google Scholar * Liu, G. Q. et al. ISG15-dependent activation of the sensor MDA5 is antagonized
by the SARS-CoV-2 papain-like protease to evade host innate immunity. _Nat. Microbiol._ 6, 467–478 (2021). CAS PubMed PubMed Central Google Scholar * Moustaqil, M. et al. SARS-CoV-2
proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. _Emerg. Microbes Infect._ 10,
178–195 (2021). CAS PubMed PubMed Central Google Scholar * Shin, D. et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. _Nature_ 587, 657–662 (2020). CAS
PubMed ADS PubMed Central Google Scholar * Zhang, S., Wang, J. & Cheng, G. Protease cleavage of RNF20 facilitates coronavirus replication via stabilization of SREBP1. _Proc. Natl
Acad. Sci. USA_ 118, e2107108118 (2021). CAS PubMed PubMed Central Google Scholar * Miao, G. et al. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the
SNARE complex required for autolysosome formation. _Dev. Cell_ 56, 427–442.e5 (2021). CAS PubMed Google Scholar * Sui, L. et al. SARS-CoV-2 membrane protein inhibits type i interferon
production through ubiquitin-mediated degradation of TBK1. _Front. Immunol._ 12, 662989 (2021). CAS PubMed PubMed Central Google Scholar * Ren, Y. et al. The ORF3a protein of SARS-CoV-2
induces apoptosis in cells. _Cell Mol. Immunol._ 17, 881–883 (2020). CAS PubMed PubMed Central Google Scholar * Zhang, Y. et al. The SARS-CoV-2 protein ORF3a inhibits fusion of
autophagosomes with lysosomes. _Cell Discov._ 7, 31 (2021). MathSciNet CAS PubMed PubMed Central Google Scholar * Ashour, H. M., Elkhatib, W. F., Md. Rahman, M. & Elshabrawy, H. A.
Insights into the recent 2019 novel coronavirus (SARS-CoV-2) in light of past human coronavirus outbreaks. _Pathogens_ 9, 186 (2020). CAS PubMed PubMed Central Google Scholar * Shang, J.
et al. Compositional diversity and evolutionary pattern of coronavirus accessory proteins. _Brief. Bioinform_ 22, 1267–1278 (2021). CAS PubMed Google Scholar * Frieman, M. et al. Severe
acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. _J. Virol._ 81, 9812–9824
(2007). CAS PubMed PubMed Central Google Scholar * Addetia, A. et al. SARS-CoV-2 ORF6 disrupts bidirectional nucleocytoplasmic transport through interactions with Rae1 and Nup98. _mBio_
12, e00065–e00121 (2021). CAS PubMed PubMed Central Google Scholar * Kato, K. et al. Overexpression of SARS-CoV-2 protein ORF6 dislocates RAE1 and NUP98 from the nuclear pore complex.
_Biochem. Biophys. Res. Commun._ 536, 59–66 (2021). CAS PubMed Google Scholar * Kimura, I. et al. Sarbecovirus ORF6 proteins hamper induction of interferon signaling. _Cell Rep._ 34,
108916 (2021). CAS PubMed PubMed Central Google Scholar * Miorin, L. et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. _Proc. Natl
Acad. Sci. USA_ 117, 28344–28354 (2020). CAS PubMed ADS PubMed Central Google Scholar * Miyamoto, Y. et al. SARS-CoV-2 ORF6 disrupts nucleocytoplasmic trafficking to advance viral
replication. _Commun. Biol._ 5, 483 (2022). CAS PubMed PubMed Central Google Scholar * Kehrer, T. et al. Impact of SARS-CoV-2 ORF6 and its variant polymorphisms on host responses and
viral pathogenesis. _Cell Host Microbe_ 31, 1668–1684.e12 (2023). CAS PubMed Google Scholar * Zhang, K. et al. Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit
host gene expression. _Sci. Adv._ 7, eabe7386 (2021). MathSciNet CAS PubMed ADS PubMed Central Google Scholar * Schubert, K. et al. Universal features of Nsp1-mediated translational
shutdown by coronaviruses. _Mol. Cell_ 83, 3546–3557.e8 (2023). CAS PubMed Google Scholar * Baggen, J., Vanstreels, E., Jansen, S. & Daelemans, D. Cellular host factors for SARS-CoV-2
infection. _Nat. Microbiol._ 6, 1219–1232 (2021). CAS PubMed Google Scholar * Baggen, J. et al. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2.
_Nat. Genet._ 53, 435–444 (2021). CAS PubMed Google Scholar * Daniloski, Z. et al. Identification of required host factors for SARS-CoV-2 infection in human cells. _Cell_ 184, 92–105.e16
(2021). CAS PubMed Google Scholar * Wang, R. et al. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. _Cell_ 184, 106–119.e14 (2021). CAS PubMed
Google Scholar * Schneider, W. M. et al. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. _Cell_ 184, 120–132.e14 (2021). CAS PubMed Google Scholar *
Baggen, J. et al. TMEM106B is a receptor mediating ACE2-independent SARS-CoV-2 cell entry. _Cell_ 186, 3427–3442.e22 (2023). CAS PubMed PubMed Central Google Scholar * Rebendenne, A. et
al. Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal HCoVs. _Nat. Genet._ 54, 1090–1102 (2022). CAS PubMed Google Scholar *
Biering, S. B. et al. Genome-wide bidirectional CRISPR screens identify mucins as host factors modulating SARS-CoV-2 infection. _Nat. Genet._ 54, 1078–1089 (2022). CAS PubMed PubMed
Central Google Scholar * Zhu, Y. et al. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. _Nat. Commun._ 12, 961 (2021). CAS PubMed ADS PubMed Central
Google Scholar * Grodzki, M. et al. Genome-scale CRISPR screens identify host factors that promote human coronavirus infection. _Genome Med._ 14, 10 (2022). CAS PubMed PubMed Central
Google Scholar * Hoffmann, H. H. et al. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. _Cell Host Microbe_ 29,
267–280.e5 (2021). CAS PubMed Google Scholar * Burkard, C. et al. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. _PLoS Pathog._ 10,
e1004502 (2014). PubMed PubMed Central Google Scholar * Schmidt, N. et al. The SARS-CoV-2 RNA–protein interactome in infected human cells. _Nat. Microbiol._ 6, 339–353 (2020). THE AUTHORS
PROVIDE INSIGHT INTO CELLULAR HOST FACTORS THAT DIRECTLY BIND SARS-COV-2 RNA IN INFECTED HUMAN CELLS AND COMPARE/EXPLAIN PREVIOUSLY DESCRIBED FUNCTIONS. PubMed PubMed Central Google
Scholar * May, D. G. et al. A BioID-derived proximity interactome for SARS-CoV-2 proteins. _Viruses_ 14, 611 (2022). CAS PubMed PubMed Central Google Scholar * Ugalde, A. P. et al.
Autophagy‐linked plasma and lysosomal membrane protein PLAC8 is a key host factor for SARS‐CoV‐2 entry into human cells. _EMBO J._ 41, e110727 (2022). CAS PubMed PubMed Central Google
Scholar * mac Kain, A. et al. Identification of DAXX as a restriction factor of SARS-CoV-2 through a CRISPR/Cas9 screen. _Nat. Commun._ 13, 2442 (2022). ADS Google Scholar * Brown, M. S.
& Goldstein, J. L. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. _Proc. Natl Acad. Sci. USA_ 96, 11041–11048 (1999). CAS PubMed ADS
PubMed Central Google Scholar * Kratzel, A. et al. A genome-wide CRISPR screen identifies interactors of the autophagy pathway as conserved coronavirus targets. _PLoS Biol._ 19, e3001490
(2021). CAS PubMed PubMed Central Google Scholar * Li, Y. E. et al. TMEM41B and VMP1 are scramblases and regulate the distribution of cholesterol and phosphatidylserine. _J. Cell Biol._
220, e202103105 (2021). CAS PubMed PubMed Central Google Scholar * Lee, S. et al. The SARS-CoV-2 RNA interactome. _Mol. Cell_ 81, 2838–2850.e6 (2021). CAS PubMed PubMed Central Google
Scholar * Labeau, A. et al. Characterization and functional interrogation of the SARS-CoV-2 RNA interactome. _Cell Rep._ 39, 110744 (2022). CAS PubMed PubMed Central Google Scholar *
Flynn, R. A. et al. Discovery and functional interrogation of SARS-CoV-2 RNA-host protein interactions. _Cell_ 184, 2394–2411.e16 (2021). CAS PubMed PubMed Central Google Scholar *
Kamel, W. et al. Global analysis of protein–RNA interactions in SARS-CoV-2-infected cells reveals key regulators of infection. _Mol. Cell_ 81, 2851–2867.e7 (2021). CAS PubMed PubMed
Central Google Scholar * Chen, Z. et al. Interactomes of SARS-CoV-2 and human coronaviruses reveal host factors potentially affecting pathogenesis. _EMBO J._ 40, e107776 (2021). CAS
PubMed PubMed Central Google Scholar * Kruse, T. et al. Large scale discovery of coronavirus-host factor protein interaction motifs reveals SARS-CoV-2 specific mechanisms and
vulnerabilities. _Nat. Commun._ 12, 6761 (2021). CAS PubMed ADS PubMed Central Google Scholar * Zheng, Z.-Q., Wang, S.-Y., Xu, Z.-S., Fu, Y.-Z. & Wang, Y.-Y. SARS-CoV-2 nucleocapsid
protein impairs stress granule formation to promote viral replication. _Cell Discov._ 7, 38 (2021). CAS PubMed PubMed Central Google Scholar * Kumar, A. et al. SARS-CoV-2 nonstructural
protein 1 inhibits the interferon response by causing depletion of key host signaling factors. _J. Virol._ 95, e0026621 (2021). PubMed Google Scholar * Xu, Z. et al. SARS-CoV-2 impairs
interferon production via NSP2-induced repression of mRNA translation. _Proc. Natl Acad. Sci. USA_ 119, e2204539119 (2022). CAS PubMed PubMed Central Google Scholar * Alhammad, Y. M. O.
et al. The SARS-CoV-2 conserved macrodomain is a mono-ADP-ribosylhydrolase. _J. Virol._ 95, e01969–e02020 (2021). CAS PubMed PubMed Central Google Scholar * Liu, Y. et al. SARS-CoV-2
Nsp5 demonstrates two distinct mechanisms targeting RIG-I and MAVS to evade the innate immune response. _mBio_ 12, e0233521 (2021). PubMed Google Scholar * Zheng, Y. et al. SARS-CoV-2 NSP5
and N protein counteract the RIG-I signaling pathway by suppressing the formation of stress granules. _Signal. Transduct. Target. Ther._ 7, 22 (2022). Google Scholar * Bhardwaj, T. et al.
Amyloidogenic proteins in the SARS-CoV and SARS-CoV-2 proteomes. _Nat. Commun._ 14, 945 (2023). CAS PubMed ADS PubMed Central Google Scholar * Fung, S. Y. et al. SARS-CoV-2 NSP13
helicase suppresses interferon signaling by perturbing JAK1 phosphorylation of STAT1. _Cell Biosci._ 12, 36 (2022). CAS PubMed PubMed Central Google Scholar * Hsu, J. C.-C.,
Laurent-Rolle, M., Pawlak, J. B., Wilen, C. B. & Cresswell, P. Translational shutdown and evasion of the innate immune response by SARS-CoV-2 NSP14 protein. _Proc. Natl Acad. Sci. USA_
118, e2101161118 (2021). PubMed PubMed Central Google Scholar * Ricardo-Lax, I. et al. Replication and single-cycle delivery of SARS-CoV-2 replicons. _Science_ 374, 1099–1106 (2021). CAS
PubMed ADS PubMed Central Google Scholar * Benton, D. J. et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. _Nature_ 588, 327–330 (2020). CAS
PubMed ADS PubMed Central Google Scholar * Freitas, R. S., Crum, T. F. & Parvatiyar, K. SARS-CoV-2 spike antagonizes innate antiviral immunity by targeting interferon regulatory
factor 3. _Front. Cell. Infect. Microbiol._ 11, 789462 (2022). PubMed PubMed Central Google Scholar * Zhang, Q. et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
membrane (M) and spike (S) proteins antagonize host type I interferon response. _Front. Cell Infect. Microbiol._ 11, 1242 (2021). Google Scholar * Fu, Y.-Z. et al. SARS-CoV-2 membrane
glycoprotein M antagonizes the MAVS-mediated innate antiviral response. _Cell Mol. Immunol._ 18, 613–620 (2021). CAS PubMed Google Scholar * Luo, L. et al. SARS-CoV-2 nucleocapsid protein
phase separates with G3BPs to disassemble stress granules and facilitate viral production. _Sci. Bull._ 66, 1194–1204 (2021). CAS Google Scholar * Wang, S. et al. Targeting liquid–liquid
phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. _Nat. Cell Biol._ 23, 718–732 (2021). CAS PubMed Google Scholar * Gori
Savellini, G., Anichini, G., Gandolfo, C. & Cusi, M. G. SARS-CoV-2 N protein targets TRIM25-mediated RIG-I activation to suppress innate immunity. _Viruses_ 13, 1439 (2021). CAS PubMed
PubMed Central Google Scholar * Oh, S. J. & Shin, O. S. SARS-CoV-2 nucleocapsid protein targets RIG-I-like receptor pathways to inhibit the induction of interferon response. _Cells_
10, 530 (2021). CAS PubMed PubMed Central Google Scholar * Chen, K. et al. SARS-CoV-2 nucleocapsid protein interacts with RIG-I and represses RIG-mediated IFN-β production. _Viruses_ 13,
47 (2020). PubMed PubMed Central Google Scholar * Mu, J. et al. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of
STAT1 and STAT2. _Cell Discov._ 6, 65 (2020). CAS PubMed PubMed Central Google Scholar * Kern, D. M. et al. Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. _Nat. Struct. Mol.
Biol._ 28, 573–582 (2021). CAS PubMed PubMed Central Google Scholar * Wang, R. et al. ORF3a protein of severe acute respiratory syndrome coronavirus 2 inhibits interferon-activated Janus
kinase/signal transducer and activator of transcription signaling via elevating suppressor of cytokine signaling 1. _Front. Microbiol._ 12, 2871 (2021). Google Scholar * Qu, Y. et al.
ORF3a-mediated incomplete autophagy facilitates severe acute respiratory syndrome coronavirus-2 replication. _Front. Cell Dev. Biol._ 9, 716208 (2021). PubMed PubMed Central Google Scholar
* Stewart, H. et al. The SARS-CoV-2 protein ORF3c is a mitochondrial modulator of innate immunity. _iScience_ 26, 108080 (2023). PubMed ADS PubMed Central Google Scholar * Cao, Z. et
al. Ubiquitination of SARS-CoV-2 ORF7a promotes antagonism of interferon response. _Cell Mol. Immunol._ 18, 746–748 (2021). CAS PubMed Google Scholar * Wu, J. et al. SARS-CoV-2 ORF9b
inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. _Cell Rep._ 34, 108761 (2021). CAS PubMed PubMed Central Google Scholar * Gao, X. et al.
Crystal structure of SARS-CoV-2 Orf9b in complex with human TOM70 suggests unusual virus–host interactions. _Nat. Commun._ 12, 2843 (2021). CAS PubMed ADS PubMed Central Google Scholar
* Han, L. et al. SARS‐CoV‐2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG‐I/MDA‐5–MAVS, TLR3–TRIF, and cGAS–STING signaling pathways. _J. Med.
Virol._ 93, 5376–5389 (2021). CAS PubMed PubMed Central Google Scholar * Focosi, D. & Maggi, F. Recombination in coronaviruses, with a focus on SARS-CoV-2. _Viruses_ 14, 1239 (2022).
CAS PubMed PubMed Central Google Scholar * Amoutzias, G. D. et al. The remarkable evolutionary plasticity of coronaviruses by mutation and recombination: insights for the COVID-19
pandemic and the future evolutionary paths of SARS-CoV-2. _Viruses_ 14, 78 (2022). CAS PubMed PubMed Central Google Scholar * Nikolaidis, M., Markoulatos, P., van de Peer, Y., Oliver, S.
G. & Amoutzias, G. D. The neighborhood of the spike gene is a hotspot for modular intertypic homologous and nonhomologous recombination in coronavirus genomes. _Mol. Biol. Evol._ 39,
msab292 (2022). CAS PubMed Google Scholar * Müller, N. F., Kistler, K. E. & Bedford, T. A Bayesian approach to infer recombination patterns in coronaviruses. _Nat. Commun._ 13, 4186
(2022). PubMed ADS PubMed Central Google Scholar * Gribble, J. et al. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. _PLoS Pathog._ 17, e1009226
(2021). THIS ARTICLE PROVIDES A FUNCTIONAL LINK BETWEEN THE PROOFREADING EXORIBONUCLEASE AND CORONAVIRUS RECOMBINATION. CAS PubMed PubMed Central Google Scholar * Turakhia, Y. et al.
Pandemic-scale phylogenomics reveals the SARS-CoV-2 recombination landscape. _Nature_ 609, 994–997 (2022). CAS PubMed ADS PubMed Central Google Scholar * Threat assessment brief:
implications for the EU/EEA of the spread of the SARS-CoV-2 Omicron XBB.1.5 sub-lineage. _ECDC_
https://www.ecdc.europa.eu/en/publications-data/covid-19-threat-assessment-brief-implications-spread-omicron-xbb (2023). * Ogando, N. S. et al. SARS-coronavirus-2 replication in Vero E6
cells: replication kinetics, rapid adaptation and cytopathology. _J. Gen. Virol._ 101, 925–940 (2020). CAS PubMed PubMed Central Google Scholar * Sasaki, M. et al. SARS-CoV-2 variants
with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. _PLoS Pathog._ 17, e1009233 (2021). THE IMPORTANCE OF THE S1/S2 FURIN CLEAVAGE
SITE AND ITS RAPID LOSS AFTER IN VITRO VIRUS PASSAGES IS DEMONSTRATED. CAS PubMed PubMed Central Google Scholar * Hou, Y. J. et al. SARS-CoV-2 D614G variant exhibits efficient
replication ex vivo and transmission in vivo. _Science_ 370, 1464–1468 (2020). CAS PubMed ADS PubMed Central Google Scholar * Han, Y., Yang, L., Lacko, L. A. & Chen, S. Human
organoid models to study SARS-CoV-2 infection. _Nat. Methods_ 19, 418–428 (2022). CAS PubMed Google Scholar * Hui, K. P. Y. et al. SARS-CoV-2 Omicron variant replication in human bronchus
and lung ex vivo. _Nature_ 603, 715–720 (2022). CAS PubMed ADS Google Scholar * Zhou, B. et al. SARS-CoV-2 spike D614G change enhances replication and transmission. _Nature_ 592,
122–127 (2021). CAS PubMed ADS Google Scholar * Chu, H., Chan, J. F. W. & Yuen, K. Y. Animal models in SARS-CoV-2 research. _Nat. Methods_ 19, 392–394 (2022). CAS PubMed Google
Scholar * Dinnon, K. H. et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. _Nature_ 586, 560–566 (2020). THESE AUTHORS CREATED A MOUSE-ADAPTED SARS-COV-2, WHICH
MODELS COVID-19 PATHOGENESIS IN MICE AND SERVES AS AN IMPORTANT MODEL SYSTEM FOR IN VIVO EVALUATION OF THERAPEUTICS AND VACCINES. PubMed ADS PubMed Central Google Scholar * Corbett, K.
S. et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. _N. Engl. J. Med._ 383, 1544–1555 (2020). CAS PubMed Google Scholar Download references
ACKNOWLEDGEMENTS The authors thank all our colleagues in the field of coronaviruses, who have helped to increase our knowledge of this virus family since its discovery. Owing to the scope
and focus of this manuscript, they have primarily focused on recent publications working with SARS-CoV-2; however, we appreciate that many of these insights are built on a strong foundation
of research on previous coronaviruses during the past decades. Work in the authors’ laboratory was supported by the Swiss National Science Foundation (grants 310030B_201278 and NCCR ‘RNA
& Disease’ grant 205601). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Institute of Virology and Immunology, Bern and Mittelhäusern, Bern, Switzerland Silvio Steiner, Annika Kratzel, G.
Tuba Barut, Reto M. Lang, Etori Aguiar Moreira, Lisa Thomann, Jenna N. Kelly & Volker Thiel * Department of Infectious Diseases and Pathobiology, Vetsuisse Faculty, University of Bern,
Bern, Switzerland Silvio Steiner, Annika Kratzel, G. Tuba Barut, Reto M. Lang, Etori Aguiar Moreira, Lisa Thomann, Jenna N. Kelly & Volker Thiel * Graduate School for Cellular and
Biomedical Sciences, University of Bern, Bern, Switzerland Reto M. Lang * Multidisciplinary Center for Infectious Diseases, University of Bern, Bern, Switzerland Jenna N. Kelly & Volker
Thiel * European Virus Bioinformatics Center, Jena, Germany Jenna N. Kelly & Volker Thiel Authors * Silvio Steiner View author publications You can also search for this author inPubMed
Google Scholar * Annika Kratzel View author publications You can also search for this author inPubMed Google Scholar * G. Tuba Barut View author publications You can also search for this
author inPubMed Google Scholar * Reto M. Lang View author publications You can also search for this author inPubMed Google Scholar * Etori Aguiar Moreira View author publications You can
also search for this author inPubMed Google Scholar * Lisa Thomann View author publications You can also search for this author inPubMed Google Scholar * Jenna N. Kelly View author
publications You can also search for this author inPubMed Google Scholar * Volker Thiel View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS
S.S., A.K., G.T.B., R.M.L., J.N.K., E.A.M. and V.T. researched the data for the article. S.S., A.K., G.T.B., R.M.L., J.N.K. and V.T. wrote the manuscript. S.S., A.K., G.T.B. and R.M.L.
prepared the figures, under the supervision of V.T. L.T. contributed to writing and revising the text. CORRESPONDING AUTHOR Correspondence to Volker Thiel. ETHICS DECLARATIONS COMPETING
INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Reviews Microbiology_ thanks Luis Enjuanes, who co-reviewed with Sonia Zuñiga; Timothy
Sheahan, who co-reviewed with Meghan Diefenbacher; and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. ADDITIONAL INFORMATION PUBLISHER’S NOTE
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. GLOSSARY * _Cis_-acting elements RNA elements found in RNA viruses that
regulate processes such as viral transcription, replication, packaging or expression of genes on the same RNA. * Double-membrane vesicles Virus-induced organelles that are delimited by two
membrane bilayers and dedicated to replication or transcription of the viral RNA. * Internal ribosomal entry sites RNA elements that allow binding of ribosomes and initiation of translation
in a cap-independent manner. * Lipid droplets Cellular lipid depositories that regulate lipid storage and metabolism. * Liquid–liquid phase separation A process that establishes two distinct
phases from a homogeneous solution; in the context of macromolecules, it is often related to the formation of membrane-less condensates of proteins and RNA. * Monocistronic Refers to
messenger RNA molecules that encode a single gene product. * Polycistronic Refers to mRNA molecules that encode more than one gene product. * Viroporin A viral protein with hydrophobic
region that can assemble into ion channels or molecular pores on membranes. RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to
this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the
terms of such publishing agreement and applicable law. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Steiner, S., Kratzel, A., Barut, G.T. _et al._ SARS-CoV-2 biology and
host interactions. _Nat Rev Microbiol_ 22, 206–225 (2024). https://doi.org/10.1038/s41579-023-01003-z Download citation * Accepted: 01 December 2023 * Published: 15 January 2024 * Issue
Date: April 2024 * DOI: https://doi.org/10.1038/s41579-023-01003-z SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a
shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative