Play all audios:
Lytic polysaccharide monooxygenases (LPMOs) are copper-containing enzymes that play a key role in the oxidative degradation of various biopolymers such as cellulose and chitin. While hunting
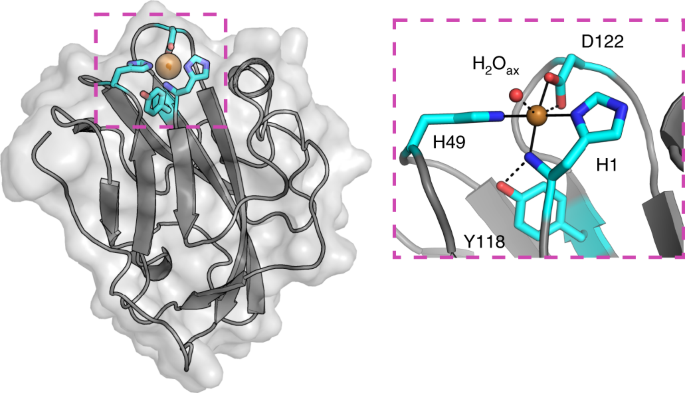
for new LPMOs, we identified a new family of proteins, defined here as X325, in various fungal lineages. The three-dimensional structure of X325 revealed an overall LPMO fold and a His
brace with an additional Asp ligand to Cu(II). Although LPMO-type activity of X325 members was initially expected, we demonstrated that X325 members do not perform oxidative cleavage of
polysaccharides, establishing that X325s are not LPMOs. Investigations of the biological role of X325 in the ectomycorrhizal fungus Laccaria bicolor revealed exposure of the X325 protein at
the interface between fungal hyphae and tree rootlet cells. Our results provide insights into a family of copper-containing proteins, which is widespread in the fungal kingdom and is
evolutionarily related to LPMOs, but has diverged to biological functions other than polysaccharide degradation.
LaX325 nucleotide sequence was deposited in GenBank under accession number MK088083. The X-ray structures of LaX325 were deposited in the Protein Data Bank under accession numbers 6IBH, 6IBI
and 6IBJ. Raw EPR data are available on request through the Research Data York (https://doi.org/10.15124/a034974e-2782-415e-8b02-2b6e4098760e).
A.L. was funded by a Marie Curie Individual Fellowship within the Horizon 2020 Research and Innovation Framework Programme (748758). The Danish Ministry of Higher Education and Science
through the Instrument Center DANSCATT funded travel to synchrotrons. We would also like to acknowledge MAX-IV, Lund, Sweden, the ESRF, Grenoble, France and DESY, Hamburg, Germany for
synchrotron beamtime and related assistance. We thank J.-C. Poulsen for technical assistance; A. Kohler and D. Thiele for helpful discussions; F. Chaspoul for assistance with ICP-MS analyses
and R. Balestrini (Institute for Sustainable Plant Protection, Italy) for providing gold-labeled WGA lectin. L.L.L. and T.T. thank the Novo Nordisk Foundation for funding through grant
NF17SA0027704. K.E.H.F. thanks the Carlsberg Foundation through an Internationalization Postdoc Fellowship (grants CF16-0673 and CF17-0533) and the EU, framework of the Marie Curie FP7
COFUND People Programme (AgreenSkills+ fellowship 609398) for financial support. L.L.L., T.T. and K.E.H.F. are members of ISBUC Integrative Structural Biology at the University of Copenhagen
(https://isbuc.ku.dk/). K.S.J. thanks the Novo Nordisk Foundation for funding through grant NNF17SA0027704. P.H.W. and L.C. thank the UK Biotechnology and Biological Sciences Research
Council (BB/L021633/1) for funding. F.M. is funded by the French National Research Agency through the Laboratory of Excellence Advanced Research on the Biology of Tree and Forest Ecosystems
(grant ANR-11-LABX 0002 01). A.Z. thanks the French Embassy in Greece, together with the French Ministry of Higher Education, Research and Innovation, for the scholarship ‘Séjour
scientifique de haut niveau’ (SSHN). The electron microscopy experiments were performed on the PiCSL-FBI core facility (IBDM, Marseille), member of the France-BioImaging national research
infrastructure (ANR-10-INBS-04).
INRA, Biodiversité et Biotechnologie Fongiques (BBF), UMR1163, Aix Marseille Université, Marseille, France
Aurore Labourel, Kristian E. H. Frandsen, Sacha Grisel, Mireille Haon, David Navarro, Marie-Noëlle Rosso, Bastien Bissaro, Anastasia Zerva & Jean-Guy Berrin
Biological Chemistry Section, Department of Chemistry, University of Copenhagen, Copenhagen, Denmark
INRA, UMR1136, Interactions Arbres/Microorganismes, Laboratoire d’Excellence ARBRE, Centre INRA-Lorraine, Université de Lorraine, Champenoux, France
INRA, Unité de Recherche Biopolymères Interactions Assemblages (BIA), Nantes, France
Department of Geosciences and Natural Resource Management, University of Copenhagen, Copenhagen, Denmark
Biotechnology Laboratory, School of Chemical Engineering, National Technical University of Athens, Zografou Campus, Athens, Greece
INRA, USC1408 Architecture et Fonction des Macromolécules Biologiques (AFMB), Marseille, France
Architecture et Fonction des Macromolécules Biologiques (AFMB), CNRS, Aix Marseille Université, Marseille, France
Department of Biological Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
A.L., D.N. and M.-N.R. identified the new proteins. A.L. and B.H. performed bioinformatic analyses. A.L., S.G. and M.H. performed recombinant protein production and purification. T.T. and
K.E.H.F. crystallized LaX325, determined and analyzed the X-ray crystal structure. T.T. collected X-ray data. K.E.H.F. made relevant figures and tables. L.L.L. directed the crystallographic
studies. K.E.H.F., K.S.J. and L.L.L. analyzed the structure. K.E.H.F. and L.L.L. drafted relevant parts of the manuscript. A.L., B.B. and A.Z. performed enzyme assays. M.F. and D.R.
performed mass spectrometry analyses. L.C. and P.H.W. conceived and carried out the EPR study. A.L. and F.Z. performed confocal microscopy under the supervision of F.M. A.L. and N.B.
performed transmission electron microscopy. J.-G.B. coordinated the work. A.L. and J.-G.B. organized the data and drafted the manuscript. All authors made comments on the manuscript and
approved the final version.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Anyone you share the following link with will be able to read this content: