Play all audios:
ABSTRACT Compact CRISPR-Cas systems offer versatile treatment options for genetic disorders, but their application is often limited by modest gene-editing activity. Here we present
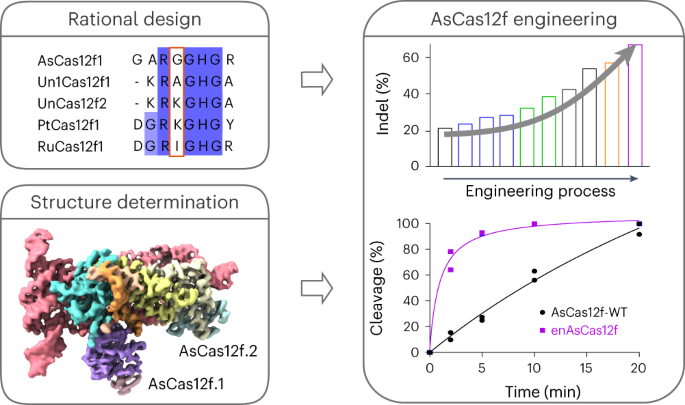
enAsCas12f, an engineered RNA-guided DNA endonuclease up to 11.3-fold more potent than its parent protein, AsCas12f, and one-third of the size of SpCas9. enAsCas12f shows higher DNA cleavage
activity than wild-type AsCas12f in vitro and functions broadly in human cells, delivering up to 69.8% insertions and deletions at user-specified genomic loci. Minimal off-target editing is
observed with enAsCas12f, suggesting that boosted on-target activity does not impair genome-wide specificity. We determine the cryo-electron microscopy (cryo-EM) structure of the
AsCas12f–sgRNA–DNA complex at a resolution of 2.9 Å, which reveals dimerization-mediated substrate recognition and cleavage. Structure-guided single guide RNA (sgRNA) engineering leads to
sgRNA-v2, which is 33% shorter than the full-length sgRNA, but with on par activity. Together, the engineered hypercompact AsCas12f system enables robust and faithful gene editing in
mammalian cells. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access
Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 print
issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to
local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT
BEING VIEWED BY OTHERS GENE EDITING WITH CRISPR-CAS12A GUIDES POSSESSING RIBOSE-MODIFIED PSEUDOKNOT HANDLES Article Open access 15 November 2021 MICAS9 INCREASES LARGE SIZE GENE KNOCK-IN
RATES AND REDUCES UNDESIRABLE ON-TARGET AND OFF-TARGET INDEL EDITS Article Open access 27 November 2020 PAM-FLEXIBLE GENOME EDITING WITH AN ENGINEERED CHIMERIC CAS9 Article Open access 04
October 2023 DATA AVAILABILITY Sequencing data are available at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) under accession number GSE211600 and
the Sequence Read Archive (SRA) under accession number PRJNA962057. Cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB, https://www.ebi.ac.uk/emdb) under accession
code EMD-27801. The atomic model has been deposited to the Protein Data Bank (PDB, https://www.rcsb.org) under accession code 8DZJ. CHANGE HISTORY * _ 01 DECEMBER 2023 A Correction to this
paper has been published: https://doi.org/10.1038/s41589-023-01508-x _ REFERENCES * Mojica, F. J. M., Díez-Villaseñor, C., García-Martínez, J. & Soria, E. Intervening sequences of
regularly spaced prokaryotic repeats derive from foreign genetic elements. _J. Mol. Evol._ 60, 174–182 (2005). Article CAS PubMed Google Scholar * Barrangou, R. et al. CRISPR provides
acquired resistance against viruses in prokaryotes. _Science_ 315, 1709–1712 (2007). Article CAS PubMed Google Scholar * Gasiunas, G., Barrangou, R., Horvath, P. & Siksnys, V.
Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. _Proc. Natl Acad. Sci. USA_ 109, E2579–E2586 (2012). Article CAS PubMed PubMed
Central Google Scholar * Jinek, M. et al. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. _Science_ 337, 816–821 (2012). Article CAS PubMed PubMed
Central Google Scholar * Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. _Nat. Biotechnol._ 38,
824–844 (2020). Article CAS PubMed Google Scholar * Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without
double-stranded DNA cleavage. _Nature_ 533, 420–424 (2016). Article CAS PubMed PubMed Central Google Scholar * Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic
DNA without DNA cleavage. _Nature_ 551, 464–471 (2017). Article CAS PubMed PubMed Central Google Scholar * Anzalone, A. V. et al. Search-and-replace genome editing without double-strand
breaks or donor DNA. _Nature_ 576, 149–157 (2019). Article CAS PubMed PubMed Central Google Scholar * Nakamura, M., Gao, Y., Dominguez, A. A. & Qi, L. S. CRISPR technologies for
precise epigenome editing. _Nat. Cell Biol._ 23, 11–22 (2021). Article CAS PubMed Google Scholar * Terns, M. P. CRISPR-based technologies: impact of RNA-targeting systems. _Mol. Cell_
72, 404–412 (2018). Article CAS PubMed PubMed Central Google Scholar * Liu, X.-M., Zhou, J., Mao, Y., Ji, Q. & Qian, S.-B. Programmable RNA _N_6-methyladenosine editing by
CRISPR-Cas9 conjugates. _Nat. Chem. Biol._ 15, 865–871 (2019). Article CAS PubMed PubMed Central Google Scholar * Wilson, C., Chen, P. J., Miao, Z. & Liu, D. R. Programmable m6A
modification of cellular RNAs with a Cas13-directed methyltransferase. _Nat. Biotechnol._ 38, 1431–1440 (2020). Article CAS PubMed PubMed Central Google Scholar * Hampton, T. With first
CRISPR trials, gene editing moves toward the clinic. _JAMA_ 323, 1537–1539 (2020). Article PubMed Google Scholar * Gillmore, J. D. et al. CRISPR-Cas9 in vivo gene editing for
transthyretin amyloidosis. _N. Engl. J. Med._ 385, 493–502 (2021). Article CAS PubMed Google Scholar * Frangoul, H. et al. CRISPR-Cas9 gene editing for sickle cell disease and
β-thalassemia. _N. Engl. J. Med._ 384, 252–260 (2021). Article CAS PubMed Google Scholar * Koonin, E. V. & Makarova, K. S. Evolutionary plasticity and functional versatility of
CRISPR systems. _PLoS Biol._ 20, e3001481 (2022). Article CAS PubMed PubMed Central Google Scholar * Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. _Science_
339, 819–823 (2013). Article CAS PubMed PubMed Central Google Scholar * Mali, P. et al. RNA-guided human genome engineering via Cas9. _Science_ 339, 823–826 (2013). Article CAS PubMed
PubMed Central Google Scholar * Jinek, M. et al. RNA-programmed genome editing in human cells. _eLife_ 2, e00471 (2013). Article PubMed PubMed Central Google Scholar * Zetsche, B. et
al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. _Cell_ 163, 759–771 (2015). Article CAS PubMed PubMed Central Google Scholar * Pickar-Oliver, A. &
Gersbach, C. A. The next generation of CRISPR–Cas technologies and applications. _Nat. Rev. Mol. Cell Biol._ 20, 490–507 (2019). Article CAS PubMed PubMed Central Google Scholar * Yin,
H., Kauffman, K. J. & Anderson, D. G. Delivery technologies for genome editing. _Nat. Rev. Drug Discov._ 16, 387–399 (2017). Article CAS PubMed Google Scholar * Lino, C. A., Harper,
J. C., Carney, J. P. & Timlin, J. A. Delivering CRISPR: a review of the challenges and approaches. _Drug Deliv._ 25, 1234–1257 (2018). Article CAS PubMed PubMed Central Google
Scholar * Wright, A. V. et al. Rational design of a split-Cas9 enzyme complex. _Proc. Natl Acad. Sci. USA_ 112, 2984–2989 (2015). Article CAS PubMed PubMed Central Google Scholar *
Zetsche, B., Volz, S. E. & Zhang, F. A split-Cas9 architecture for inducible genome editing and transcription modulation. _Nat. Biotechnol._ 33, 139–142 (2015). Article CAS PubMed
Google Scholar * Nihongaki, Y., Kawano, F., Nakajima, T. & Sato, M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. _Nat. Biotechnol._ 33, 755–760 (2015). Article CAS
PubMed Google Scholar * Chew, W. L. et al. A multifunctional AAV–CRISPR–Cas9 and its host response. _Nat. Methods_ 13, 868–874 (2016). Article CAS PubMed PubMed Central Google Scholar
* Liu, J.-J. et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. _Nature_ 566, 218–223 (2019). Article CAS PubMed PubMed Central Google Scholar * Pausch, P.
et al. CRISPR-CasΦ from huge phages is a hypercompact genome editor. _Science_ 369, 333–337 (2020). Article CAS PubMed PubMed Central Google Scholar * Harrington, L. B. et al.
Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. _Science_ 362, 839–842 (2018). Article CAS PubMed PubMed Central Google Scholar * Karvelis, T. et al. PAM recognition by
miniature CRISPR–Cas12f nucleases triggers programmable double-stranded DNA target cleavage. _Nucleic Acids Res._ 48, 5016–5023 (2020). Article CAS PubMed PubMed Central Google Scholar
* Lee, H. J., Kim, H. J. & Lee, S. J. Miniature CRISPR-Cas12f1-mediated single-nucleotide microbial genome editing using 3′-truncated sgRNA. _CRISPR J._ 6, 52–61 (2023). Article CAS
PubMed PubMed Central Google Scholar * Kapitonov, V. V., Makarova, K. S. & Koonin, E. V. ISC, a novel group of bacterial and archaeal DNA transposons that encode Cas9 homologs. _J.
Bacteriol._ 198, 797–807 (2016). Article CAS PubMed Central Google Scholar * Altae-Tran, H. et al. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided
endonucleases. _Science_ 374, 57–65 (2021). Article CAS PubMed PubMed Central Google Scholar * Kato, K. et al. Structure of the IscB–ωRNA ribonucleoprotein complex, the likely ancestor
of CRISPR-Cas9. _Nat. Commun._ 13, 6719 (2022). Article CAS PubMed PubMed Central Google Scholar * Hirano, S. et al. Structure of the OMEGA nickase IsrB in complex with ωRNA and target
DNA. _Nature_ 610, 575–581 (2022). Article CAS PubMed PubMed Central Google Scholar * Karvelis, T. et al. Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease.
_Nature_ 599, 692–696 (2021). Article CAS PubMed PubMed Central Google Scholar * Schuler, G., Hu, C. & Ke, A. Structural basis for RNA-guided DNA cleavage by IscB-ωRNA and
mechanistic comparison with Cas9. _Science_ 376, 1476–1481 (2022). Article CAS PubMed PubMed Central Google Scholar * Takeda, S. N. et al. Structure of the miniature type V-F CRISPR-Cas
effector enzyme. _Mol. Cell_ 81, 558–570.e3 (2021). Article CAS PubMed Google Scholar * Xiao, R., Li, Z., Wang, S., Han, R. & Chang, L. Structural basis for substrate recognition
and cleavage by the dimerization-dependent CRISPR–Cas12f nuclease. _Nucleic Acids Res._ 49, 4120–4128 (2021). Article CAS PubMed PubMed Central Google Scholar * Kong, X. et al.
Engineered CRISPR-OsCas12f1 and RhCas12f1 with robust activities and expanded target range for genome editing. _Nat. Commun._ 14, 2046 (2023). Article CAS PubMed PubMed Central Google
Scholar * Wu, Z. et al. Programmed genome editing by a miniature CRISPR-Cas12f nuclease. _Nat. Chem. Biol._ 17, 1132–1138 (2021). Article CAS PubMed Google Scholar * Xu, X. et al.
Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. _Mol. Cell_ 81, 4333–4345.e4 (2021). Article CAS PubMed Google Scholar * Kim, D. Y. et al. Efficient
CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. _Nat. Biotechnol._ 40, 94–102 (2022). Article CAS PubMed Google Scholar * Kim,
D. Y. et al. Hypercompact adenine base editors based on a Cas12f variant guided by engineered RNA. _Nat. Chem. Biol._ 18, 1005–1013 (2022). Article CAS PubMed Google Scholar * Tsai, S.
Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. _Nat. Biotechnol._ 33, 187–197 (2015). Article CAS PubMed Google Scholar * Marcovitz, A.
& Levy, Y. Frustration in protein–DNA binding influences conformational switching and target search kinetics. _Proc. Natl Acad. Sci. USA_ 108, 17957–17962 (2011). Article CAS PubMed
PubMed Central Google Scholar * Kleinstiver, B. P. et al. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing.
_Nat. Biotechnol._ 37, 276–282 (2019). Article CAS PubMed PubMed Central Google Scholar * Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis.
_Nat. Biotechnol._ 37, 224–226 (2019). Article CAS PubMed PubMed Central Google Scholar * Xin, C. et al. Comprehensive assessment of miniature CRISPR-Cas12f nucleases for gene
disruption. _Nat. Commun._ 13, 5623 (2022). Article CAS PubMed PubMed Central Google Scholar * Zhang, S. et al. TadA reprogramming to generate potent miniature base editors with high
precision. _Nat. Commun._ 14, 413 (2023). Article CAS PubMed PubMed Central Google Scholar * Chavez, A. et al. Highly efficient Cas9-mediated transcriptional programming. _Nat. Methods_
12, 326–328 (2015). Article CAS PubMed PubMed Central Google Scholar * Dang, Y. et al. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. _Genome Biol._ 16, 280
(2015). Article PubMed PubMed Central Google Scholar * Moon, S. B., Kim, D. Y., Ko, J.-H., Kim, J.-S. & Kim, Y.-S. Improving CRISPR genome editing by engineering guide RNAs. _Trends
Biotechnol._ 37, 870–881 (2019). Article CAS PubMed Google Scholar * Kleinstiver, B. P. et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. _Nat. Biotechnol._
34, 869–874 (2016). Article CAS PubMed PubMed Central Google Scholar * Bae, S., Park, J. & Kim, J.-S. Cas-OFFinder: a fast and versatile algorithm that searches for potential
off-target sites of Cas9 RNA-guided endonucleases. _Bioinformatics_ 30, 1473–1475 (2014). Article CAS PubMed PubMed Central Google Scholar * Zheng, S. Q. et al. MotionCor2: anisotropic
correction of beam-induced motion for improved cryo-electron microscopy. _Nat. Methods_ 14, 331–332 (2017). Article CAS PubMed PubMed Central Google Scholar * Punjani, A., Rubinstein,
J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. _Nat. Methods_ 14, 290–296 (2017). Article CAS PubMed Google
Scholar * Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. _J. Struct. Biol._ 180, 519–530 (2012). Article CAS PubMed PubMed Central
Google Scholar * Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. _J. Struct. Biol._ 142, 334–347 (2003). Article
PubMed Google Scholar * Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. _Acta Crystallogr. D Biol. Crystallogr._ 60, 2126–2132 (2004). Article PubMed
Google Scholar * Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. _Nature_ 596, 583–589 (2021). Article CAS PubMed PubMed Central Google Scholar * Sato,
K., Kato, Y., Hamada, M., Akutsu, T. & Asai, K. IPknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming. _Bioinformatics_ 27, i85–i93
(2011). Article CAS PubMed PubMed Central Google Scholar * Popenda, M. et al. Automated 3D structure composition for large RNAs. _Nucleic Acids Res._ 40, e112 (2012). Article CAS
PubMed PubMed Central Google Scholar * Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. _Acta Crystallogr. D Biol. Crystallogr._ 66,
213–221 (2010). Article CAS PubMed PubMed Central Google Scholar * Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. _Protein
Sci._ 30, 70–82 (2021). Article CAS PubMed Google Scholar * Malinin, N. L. et al. Defining genome-wide CRISPR–Cas genome-editing nuclease activity with GUIDE-seq. _Nat. Protoc._ 16,
5592–5615 (2021). Article CAS PubMed PubMed Central Google Scholar * Tsai, S. Q., Topkar, V. V., Joung, J. K. & Aryee, M. J. Open-source guideseq software for analysis of GUIDE-seq
data. _Nat. Biotechnol._ 34, 483 (2016). Article PubMed Google Scholar Download references ACKNOWLEDGEMENTS We thank all members of the Tang laboratory for discussion. We thank the staff
at the University of Chicago Advanced Electron Microscopy Facility (RRID: SCR_019198) for helping with cryo-EM data collection. We thank the Research Computing Center at the University of
Chicago for providing the computing resources of the Beagle 3 HPC cluster funded by the National Institutes of Health (NIH) (S10OD028655). This work was supported by the NIH under grant
number R35GM143052 to M.Z. W.T. is supported by the Searle Scholars Program (SSP-2021-113), the Cancer Research Foundation Young Investigator Program, the American Cancer Society
(RSG-22-043-01-ET), and the David & Lucile Packard Foundation. AUTHOR INFORMATION Author notes * These authors contributed equally: Tong Wu, Chang Liu, Siyuan Zou. * Deceased: Bowei
Yang. AUTHORS AND AFFILIATIONS * Department of Chemistry, The University of Chicago, Chicago, IL, USA Tong Wu, Chang Liu, Siyuan Zou, Ruitu Lyu, Hao Yan & Weixin Tang * Institute for
Biophysical Dynamics, The University of Chicago, Chicago, IL, USA Tong Wu, Siyuan Zou, Ruitu Lyu, Bowei Yang, Hao Yan, Minglei Zhao & Weixin Tang * Department of Biochemistry and
Molecular Biology, The University of Chicago, Chicago, IL, USA Bowei Yang & Minglei Zhao Authors * Tong Wu View author publications You can also search for this author inPubMed Google
Scholar * Chang Liu View author publications You can also search for this author inPubMed Google Scholar * Siyuan Zou View author publications You can also search for this author inPubMed
Google Scholar * Ruitu Lyu View author publications You can also search for this author inPubMed Google Scholar * Bowei Yang View author publications You can also search for this author
inPubMed Google Scholar * Hao Yan View author publications You can also search for this author inPubMed Google Scholar * Minglei Zhao View author publications You can also search for this
author inPubMed Google Scholar * Weixin Tang View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS All authors designed experiments and
interpreted the data. T.W., M.Z. and W.T. conceived the project. T.W. and S.Z. performed protein engineering, gRNA engineering, cellular gene-editing and in vitro DNA cleavage assays with
help and suggestions from H.Y. C.L. and B.Y. purified proteins. C.L. collected cryo-EM data and performed data analysis. R.L. assisted with GUIDE-seq data analysis. T.W., C.L., M.Z. and W.T.
wrote the paper with input from all authors. CORRESPONDING AUTHORS Correspondence to Minglei Zhao or Weixin Tang. ETHICS DECLARATIONS COMPETING INTERESTS T.W., S.Z. and W.T. are inventors
on a US provisional patent application on enAsCas12f. T.W. is a shareholder of AccuraDX. All other authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature
Chemical Biology_ thanks Jun-Jie Liu, Hyongbum Henry Kim and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. ADDITIONAL INFORMATION PUBLISHER’S NOTE
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary
Tables 1–6 and Supplementary Figs. 1–18. REPORTING SUMMARY SUPPLEMENTARY DATA 1 Sequences of DNA oligos used to amplify genomic loci for amplicon sequencing. RIGHTS AND PERMISSIONS Springer
Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author
self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law. Reprints and permissions ABOUT THIS ARTICLE
CITE THIS ARTICLE Wu, T., Liu, C., Zou, S. _et al._ An engineered hypercompact CRISPR-Cas12f system with boosted gene-editing activity. _Nat Chem Biol_ 19, 1384–1393 (2023).
https://doi.org/10.1038/s41589-023-01380-9 Download citation * Received: 23 August 2022 * Accepted: 08 June 2023 * Published: 03 July 2023 * Issue Date: November 2023 * DOI:
https://doi.org/10.1038/s41589-023-01380-9 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative