Play all audios:
ABSTRACT Live attenuated vaccines are generally highly efficacious and often superior to inactivated vaccines, yet the underlying mechanisms of this remain largely unclear. Here we identify
recognition of microbial viability as a potent stimulus for follicular helper T cell (TFH cell) differentiation and vaccine responses. Antigen-presenting cells (APCs) distinguished viable
bacteria from dead bacteria through Toll-like receptor 8 (TLR8)-dependent detection of bacterial RNA. In contrast to dead bacteria and other TLR ligands, live bacteria, bacterial RNA and
synthetic TLR8 agonists induced a specific cytokine profile in human and porcine APCs, thereby promoting TFH cell differentiation. In domestic pigs, immunization with a live bacterial
vaccine induced robust TFH cell and antibody responses, but immunization with its heat-killed counterpart did not. Finally, a hypermorphic _TLR8_ polymorphism was associated with protective
immunity elicited by vaccination with bacillus Calmette-Guérin (BCG) in a human cohort. We have thus identified TLR8 as an important driver of TFH cell differentiation and a promising target
for TFH cell–skewing vaccine adjuvants. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through
your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to this
journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now
Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer
support SIMILAR CONTENT BEING VIEWED BY OTHERS A MOLECULAR ATLAS OF INNATE IMMUNITY TO ADJUVANTED AND LIVE ATTENUATED VACCINES, IN MICE Article Open access 27 January 2022 KINETICALLY
ACTIVATING NANOVACCINE MIMICKING MULTIDIMENSIONAL IMMUNOMODULATION OF NATURAL INFECTION FOR BROAD PROTECTION AGAINST HETEROLOGOUS VIRUSES IN ANIMAL MODELS Article Open access 25 March 2025
IL6 SUPPRESSES VACCINE RESPONSES IN NEONATES BY ENHANCING IL2 ACTIVITY ON T FOLLICULAR HELPER CELLS Article Open access 08 November 2023 REFERENCES * Barquet, N. & Domingo, P. Smallpox:
the triumph over the most terrible of the ministers of death. _Ann. Intern. Med._ 127, 635–642 (1997). Article CAS PubMed Google Scholar * Plotkin, S. A. & Plotkin, S. L. The
development of vaccines: how the past led to the future. _Nat. Rev. Microbiol._ 9, 889–893 (2011). Article CAS PubMed Google Scholar * Minor, P. D. Live attenuated vaccines: Historical
successes and current challenges. _Virology_ 479–480, 379–392 (2015). Article PubMed Google Scholar * De Gregorio, E. & Rappuoli, R. From empiricism to rational design: a personal
perspective of the evolution of vaccine development. _Nat. Rev. Immunol._ 14, 505–514 (2014). Article PubMed Google Scholar * Rauh, L. W. & Schmidt, R. Measles immunization with
killed virus vaccine. Serum antibody titers and experience with exposure to measles epidemic. _Am. J. Dis. Child._ 109, 232–237 (1965). * Blander, J. M. & Sander, L. E. Beyond pattern
recognition: five immune checkpoints for scaling the microbial threat. _Nat. Rev. Immunol._ 12, 215–225 (2012). Article CAS PubMed Google Scholar * Sander, L. E. et al. Detection of
prokaryotic mRNA signifies microbial viability and promotes immunity. _Nature_ 474, 385–389 (2011). Article CAS PubMed PubMed Central Google Scholar * Iwasaki, A. & Medzhitov, R.
Control of adaptive immunity by the innate immune system. _Nat. Immunol._ 16, 343–353 (2015). Article CAS PubMed PubMed Central Google Scholar * Crotty, S. T follicular helper cell
differentiation, function, and roles in disease. _Immunity_ 41, 529–542 (2014). Article CAS PubMed PubMed Central Google Scholar * Vinuesa, C. G., Linterman, M. A., Yu, D. &
MacLennan, I. C. Follicular helper T cells. _Annu. Rev. Immunol._ 34, 335–368 (2016). Article CAS PubMed Google Scholar * Johnston, R. J. et al. Bcl6 and Blimp-1 are reciprocal and
antagonistic regulators of T follicular helper cell differentiation. _Science_ 325, 1006–1010 (2009). Article CAS PubMed PubMed Central Google Scholar * Nurieva, R. I. et al. Bcl6
mediates the development of T follicular helper cells. _Science_ 325, 1001–1005 (2009). Article CAS PubMed PubMed Central Google Scholar * Linterman, M. A. et al. IL-21 acts directly on
B cells to regulate Bcl-6 expression and germinal center responses. _J. Exp. Med._ 207, 353–363 (2010). Article CAS PubMed PubMed Central Google Scholar * Weber, J. P. et al. ICOS
maintains the T follicular helper cell phenotype by down-regulating Kruppel-like factor 2. _J. Exp. Med._ 212, 217–233 (2015). Article CAS PubMed PubMed Central Google Scholar *
Breitfeld, D. et al. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. _J. Exp. Med._ 192, 1545–1552 (2000).
Article CAS PubMed PubMed Central Google Scholar * Schaerli, P. et al. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. _J. Exp. Med._
192, 1553–1562 (2000). Article CAS PubMed PubMed Central Google Scholar * Ueno, H., Banchereau, J. & Vinuesa, C. G. Pathophysiology of T follicular helper cells in humans and mice.
_Nat. Immunol._ 16, 142–152 (2015). Article CAS PubMed PubMed Central Google Scholar * Cucak, H., Yrlid, U., Reizis, B., Kalinke, U. & Johansson-Lindbom, B. Type I interferon
signaling in dendritic cells stimulates the development of lymph-node-resident T follicular helper cells. _Immunity_ 31, 491–501 (2009). Article CAS PubMed Google Scholar * Batten, M. et
al. IL-27 supports germinal center function by enhancing IL-21 production and the function of T follicular helper cells. _J. Exp. Med._ 207, 2895–2906 (2010). Article CAS PubMed PubMed
Central Google Scholar * Schmitt, N. et al. The cytokine TGF-β co-opts signaling via STAT3-STAT4 to promote the differentiation of human TFH cells. _Nat. Immunol._ 15, 856–865 (2014).
Article PubMed PubMed Central Google Scholar * Jacquemin, C. et al. OX40 ligand contributes to human lupus pathogenesis by promoting T follicular helper response. _Immunity_ 42,
1159–1170 (2015). Article CAS PubMed PubMed Central Google Scholar * Eigenbrod, T., Pelka, K., Latz, E., Kreikemeyer, B. & Dalpke, A. H. TLR8 senses bacterial RNA in human monocytes
and plays a nonredundant role for recognition of _Streptococcus pyogenes_. _J. Immunol._ 195, 1092–1099 (2015). Article CAS PubMed Google Scholar * Bergstrom, B. et al. TLR8 senses
_Staphylococcus aureus_ RNA in human primary monocytes and macrophages and induces IFN-β production via a TAK1-IKKβ-IRF5 signaling pathway. _J. Immunol._ 195, 1100–1111 (2015). Article CAS
PubMed Google Scholar * Meurens, F., Summerfield, A., Nauwynck, H., Saif, L. & Gerdts, V. The pig: a model for human infectious diseases. _Trends Microbiol._ 20, 50–57 (2012).
Article CAS PubMed Google Scholar * Mair, K. H. et al. The porcine innate immune system: an update. _Dev. Comp. Immunol._ 45, 321–343 (2014). Article CAS PubMed Google Scholar * Zhu,
J., Lai, K., Brownile, R., Babiuk, L. A. & Mutwiri, G. K. Porcine TLR8 and TLR7 are both activated by a selective TLR7 ligand, imiquimod. _Mol. Immunol._ 45, 3238–3243 (2008). Article
CAS PubMed Google Scholar * Springer S., Lindner T., Steinbach G., Selbitz H. J. Investigation of the efficacy of a genetically-stabile live _Salmonella typhimurium_ vaccine for use in
swine. _Berl. Munch. Tierarztl. Wochenschr._ 114,342–345 (2001). * Sinkora, M., Stepanova, K. & Sinkorova, J. Different anti-CD21 antibodies can be used to discriminate developmentally
and functionally different subsets of B lymphocytes in circulation of pigs. _Dev. Comp. Immunol._ 39, 409–418 (2013). Article CAS PubMed Google Scholar * Oh, D. Y. et al. A functional
toll-like receptor 8 variant is associated with HIV disease restriction. _J. Infect. Dis._ 198, 701–709 (2008). Article CAS PubMed Google Scholar * Wang, C. H. et al. TLR7 and TLR8 gene
variations and susceptibility to hepatitis C virus infection. _PLoS One_ 6, e26235 (2011). Article CAS PubMed PubMed Central Google Scholar * Tanji, H., Ohto, U., Shibata, T., Miyake,
K. & Shimizu, T. Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands. _Science_ 339, 1426–1429 (2013). Article CAS PubMed Google Scholar *
Davila, S. et al. Genetic association and expression studies indicate a role of toll-like receptor 8 in pulmonary tuberculosis. _PLoS Genet._ 4, e1000218 (2008). Article PubMed PubMed
Central Google Scholar * Schmitt, N. et al. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. _Immunity_
31, 158–169 (2009). Article CAS PubMed PubMed Central Google Scholar * Hochrein, H. et al. Interleukin (IL)-4 is a major regulatory cytokine governing bioactive IL-12 production by
mouse and human dendritic cells. _J. Exp. Med._ 192, 823–833 (2000). Article CAS PubMed PubMed Central Google Scholar * Jones, S. C., Brahmakshatriya, V., Huston, G., Dibble, J. &
Swain, S. L. TLR-activated dendritic cells enhance the response of aged naive CD4 T cells via an IL-6-dependent mechanism. _J. Immunol._ 185, 6783–6794 (2010). Article CAS PubMed PubMed
Central Google Scholar * Chakarov, S. & Fazilleau, N. Monocyte-derived dendritic cells promote T follicular helper cell differentiation. _EMBO Mol. Med._ 6, 590–603 (2014). CAS PubMed
PubMed Central Google Scholar * Heil, F. et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. _Science_ 303, 1526–1529 (2004). Article CAS PubMed
Google Scholar * Schmitt, N. et al. IL-12 receptor beta1 deficiency alters in vivo T follicular helper cell response in humans. _Blood_ 121, 3375–3385 (2013). * Barbet, G. et al. Sensing
microbial viability through bacterial RNA augments T follicular helper cell and antibody responses. _Immunity_ (in the press). * Dowling, D. J. et al. TLR7/8 adjuvant overcomes newborn
hyporesponsiveness to pneumococcal conjugate vaccine at birth. _JCI Insight_ 2, e91020 (2017). Article PubMed PubMed Central Google Scholar * Dowling, D. J. et al. Toll-like receptor 8
agonist nanoparticles mimic immunomodulating effects of the live BCG vaccine and enhance neonatal innate and adaptive immune responses. _J. Allergy Clin. Immunol_. 140, 1339–1350 (2017). *
Fairbairn, L., Kapetanovic, R., Sester, D. P. & Hume, D. A. The mononuclear phagocyte system of the pig as a model for understanding human innate immunity and disease. _J. Leukoc. Biol._
89, 855–871 (2011). Article CAS PubMed Google Scholar * Landers, T. F., Cohen, B., Wittum, T. E. & Larson, E. L. A review of antibiotic use in food animals: perspective, policy, and
potential. _Public Health Rep._ 127, 4–22 (2012). Article PubMed PubMed Central Google Scholar * Colditz, G. A. et al. Efficacy of BCG vaccine in the prevention of tuberculosis.
Meta-analysis of the published literature. _J. Am. Med. Assoc._ 271, 698–702 (1994). * de Beaucoudrey, L. et al. Revisiting human IL-12Rβ deficiency: a survey of 141 patients from 30
countries. _Medicine (Baltimore)_ 89, 381–402 (2010). Article Google Scholar * Picard, C. et al. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. _Medicine
(Baltimore)_ 89, 403–425 (2010). Article CAS Google Scholar * Ma, C. S. et al. Functional STAT3 deficiency compromises the generation of human T follicular helper cells. _Blood_ 119,
3997–4008 (2012). Article CAS PubMed PubMed Central Google Scholar * Ma, C. S. et al. Monogenic mutations differentially affect the quantity and quality of T follicular helper cells in
patients with human primary immunodeficiencies. _J. Allergy Clin. Immunol._ 136, 993–1006 (2015). Article CAS PubMed PubMed Central Google Scholar * Rao, D. A. et al. Pathologically
expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. _Nature_ 542, 110–114 (2017). Article CAS PubMed PubMed Central Google Scholar * Lin, K. et al. MADMAX -
Management and analysis database for multiple ~omics experiments. _J. Integr. Bioinform._ 8, 160 (2011). * Dai, M. et al. Evolving gene/transcript definitions significantly alter the
interpretation of GeneChip data. _Nucleic Acids Res._ 33, e175 (2005). * Irizarry, R. A. et al. Exploration, normalization, and summaries of high density ol array probe level data.
_Biostatistics_ 4, 249–264 (2003). Article PubMed Google Scholar * Sartor, M. A. et al. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in
microarray experiments. _BMC Bioinformatics_ 7, 538 (2006). Article PubMed PubMed Central Google Scholar * Storey, J. D. & Tibshirani, R. Statistical significance for genomewide
studies. _Proc. Natl. Acad. Sci. USA_ 100, 9440–9445 (2003). Article CAS PubMed PubMed Central Google Scholar * Dittrich, N. et al. Toll-like receptor 1 variations influence
susceptibility and immune response to Mycobacterium tuberculosis. _Tuberculosis (Edinb.)_ 95, 328–335 (2015). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We thank D.
Kunkel, S. Warth and A. Linke for technical advice; the flow cytometry facility of the Berlin Brandenburg Center for Regenerative Therapies (BCRT) of the Charité Berlin; S. Springer and IDT
Biologika for assistance in planning the animal studies and for the Salmoporc STM vaccine; and R. Nifosí for critical review of the modelling analysis. Supported by the German Research
Council (DFG grant SA1940-2/1 and SFB-TR84 TP C8 to L.E.S.; SFB-TR84 TP B1 to N.S.; SFB-TR-84 TP A1/A5 to B.O.; SFB-TR84 TP Z1b to A.D.G.; DFG-GRK 1673 project to R.R.S.; and DFG-GRK 2046,
TP4 to S.H.), the European Research Council and the Federal Ministry of Education and Research (FP-7 ERA-NET / Infect-ERA consortium “HaploINFECT” / BMBF 031A402A to L.E.S., VIP + VALNEMCYS
project and InfectControl 2020, consortium Art4Fun to S.H.), the European Society of Clinical Microbiology and Infectious Diseases (ESCMID research grant to L.E.S.), the Jürgen Manchot
Foundation (Doctoral Research Fellowship to P.G., E.T.H. and S.M.V.), the Netherlands Nutrigenomics Center, Wageningen University, The Netherlands (to M.M. and M.B.), the Fritz Thyssen
Foundation (research grant to A.H.), The Danish National Research Foundation (grant no. DNRF108 to) to Research Centre for Vitamins and Vaccines (CVIVA) supporting K.J.J., and The Novo
Nordisk Foundation supporting the pig vaccination experiments at Technical University of Denmark, Federal Ministry of Education and Research. AUTHOR INFORMATION Author notes * These authors
contributed equally: Matteo Ugolini, Jenny Gerhard and Sanne Burkert. AUTHORS AND AFFILIATIONS * Department of Infectious Diseases and Pulmonary Medicine, Charité–Universitätsmedizin Berlin,
corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Berlin, Germany Matteo Ugolini, Jenny Gerhard, Philipp Georg, Sarah M. Volkers,
Elisa T. Helbig, Bastian Opitz, Florian Kurth, Norbert Suttorp & Leif E. Sander * Institute of Microbiology and Hygiene, Charité–Universitätsmedizin Berlin, corporate member of Freie
Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Berlin, Germany Sanne Burkert, Shruthi Thada, Saubashya Sur, Nickel Dittrich & Ralf R. Schumann *
Research Center for Vitamins and Vaccines, Bandim Health Project, Statens Serum Institut, Copenhagen, Denmark Kristoffer Jarlov Jensen & Christine S. Benn * Department of Biotechnology
and Biomedicine, Technical University of Denmark, Kgs Lyngby, Denmark Kristoffer Jarlov Jensen & Gregers Jungersen * Department of Veterinary Medicine, Institute of Immunology, Freie
Universität Berlin, Berlin, Germany Friederike Ebner & Susanne Hartmann * Bhagwan Mahavir Medical Research Centre, Hyderabad, India Shruthi Thada & Sumanlatha Gaddam * Department of
Veterinary Medicine, Institute of Veterinary Pathology, Freie Universität Berlin, Berlin, Germany Kristina Dietert & Achim D. Gruber * Chronic Immune Reactions, German Rheumatism
Research Centre, a Leibniz Institute, Berlin, Germany Laura Bauer & Andreas Hutloff * Institute of Immunology, Friedrich-Loeffler-Institut, Federal Research Institute for Animal Health,
Greifswald—Island of Riems, Germany Alexander Schäfer & Ulrike Blohm * German Center for Lung Research (DZL), Berlin, Germany Bastian Opitz, Norbert Suttorp & Leif E. Sander *
Department of Genetics, Osmania University, Hyderabad, India Sumanlatha Gaddam * Department of Internal Medicine and Dermatology, Division of Psychosomatic Medicine,
Charite–Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Berlin, Germany Melanie L. Conrad * Odense
Patient Data Explorative Network (OPEN), Odense University Hospital/Department of Clinical Research, University of Southern Denmark, Odense, Denmark Christine S. Benn * Nutrition, Metabolism
and Genomics Group, Division of Human Nutrition, Wageningen University, Wageningen, The Netherlands Mark V. Boekschoten & Michael Müller * Norwich Medical School, University of East
Anglia, Norwich, UK Michael Müller Authors * Matteo Ugolini View author publications You can also search for this author inPubMed Google Scholar * Jenny Gerhard View author publications You
can also search for this author inPubMed Google Scholar * Sanne Burkert View author publications You can also search for this author inPubMed Google Scholar * Kristoffer Jarlov Jensen View
author publications You can also search for this author inPubMed Google Scholar * Philipp Georg View author publications You can also search for this author inPubMed Google Scholar *
Friederike Ebner View author publications You can also search for this author inPubMed Google Scholar * Sarah M. Volkers View author publications You can also search for this author inPubMed
Google Scholar * Shruthi Thada View author publications You can also search for this author inPubMed Google Scholar * Kristina Dietert View author publications You can also search for this
author inPubMed Google Scholar * Laura Bauer View author publications You can also search for this author inPubMed Google Scholar * Alexander Schäfer View author publications You can also
search for this author inPubMed Google Scholar * Elisa T. Helbig View author publications You can also search for this author inPubMed Google Scholar * Bastian Opitz View author publications
You can also search for this author inPubMed Google Scholar * Florian Kurth View author publications You can also search for this author inPubMed Google Scholar * Saubashya Sur View author
publications You can also search for this author inPubMed Google Scholar * Nickel Dittrich View author publications You can also search for this author inPubMed Google Scholar * Sumanlatha
Gaddam View author publications You can also search for this author inPubMed Google Scholar * Melanie L. Conrad View author publications You can also search for this author inPubMed Google
Scholar * Christine S. Benn View author publications You can also search for this author inPubMed Google Scholar * Ulrike Blohm View author publications You can also search for this author
inPubMed Google Scholar * Achim D. Gruber View author publications You can also search for this author inPubMed Google Scholar * Andreas Hutloff View author publications You can also search
for this author inPubMed Google Scholar * Susanne Hartmann View author publications You can also search for this author inPubMed Google Scholar * Mark V. Boekschoten View author publications
You can also search for this author inPubMed Google Scholar * Michael Müller View author publications You can also search for this author inPubMed Google Scholar * Gregers Jungersen View
author publications You can also search for this author inPubMed Google Scholar * Ralf R. Schumann View author publications You can also search for this author inPubMed Google Scholar *
Norbert Suttorp View author publications You can also search for this author inPubMed Google Scholar * Leif E. Sander View author publications You can also search for this author inPubMed
Google Scholar CONTRIBUTIONS M.U. and J.G. designed and performed experiments; S.B. analyzed clinical data and performed experiments; K.J.J., C.S.B., G.J. and L.E.S. planned and performed
animal studies; P.G., F.E., S.M.V., S.T., K.D., L.B. and E.T.H. performed experiments; M.U., J.G., P.G., F.E., S.M.V., K.D., B.O., F.K., A.D.G., A.H., S.H., M.M., N.S. and L.E.S. analyzed
data and wrote the manuscript; S.S. performed structure prediction analysis; N.D., S.G., M.L.C. and R.R.S. designed and performed clinical studies; A.S. and U.B. provided samples and
conceptual input; M.V.B. performed transcriptome analysis; and L.E.S. conceived of the study and designed experiments. CORRESPONDING AUTHOR Correspondence to Leif E. Sander. ETHICS
DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE: Springer Nature remains neutral with regard to jurisdictional claims in
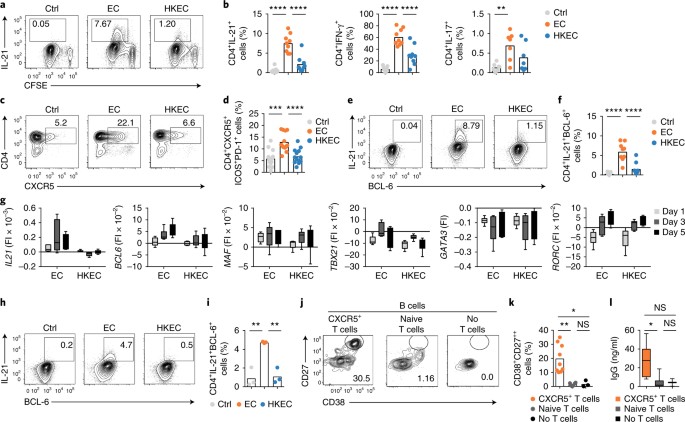
published maps and institutional affiliations. INTEGRATED SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE 1 RECOGNITION OF BACTERIAL VIABILITY INDUCES TFH CELLS. (A) Human monocytes were
stimulated with medium (ctrl), live _E. coli_ (EC) or heat killed _E. coli_ (HKEC) and co-cultured with autologous naïve CD4+ T cells in the presence of SEB. Expression of CXCR5, ICOS and
PD-1 was measured by flow cytometry. Representative FACS plots of data shown in Fig. 1d. Numbers indicate gate frequencies as ‘% of parent population’ (in black) or as ‘% of total T cells’
(in red) (B) Same samples as in Figure 1h. Flourescence intensity values (FI) corrected for background without subtraction of control FI. (C-F) CD4+ T cells were co-cultured with APC as
described in Fig. 1. T cells were sorted based on CXCR5 expression on day 5 and subsequently co-cultured with autologous naïve B cells for 7 days. Plasmablast differentiation (c, d) and Ig
class-switching (e, f) measured in cultures containing only B cells, B cells + CXCR5+ T cells, and B cells + CXCR5– T cells (n=4). One-way ANOVA with post hoc correction for multiple
comparison. Error bars are mean ± SEM (*; p<0.05, **; p<0.01). SUPPLEMENTARY FIGURE 2 IL-12 AND IN PARTS IL-1Β, BUT NOT IFN-Β ARE REQUIRED FOR EC-INDUCED TFH CELL DIFFERENTIATION. (A)
Proliferation of CD4+ T cells cultured as in Figure 4 in the presence of conditioned APC supernatants was measured by flow cytometry (CFSE dilution). (B, C) Same samples as in Figure 4a,b
respectively. IL-17A production was measured by flow cytometry (b, n=28) and ELISA (c, n=10). (D-F) CD4+ T cells were cultured and stimulated as in Figure 3d (aIL-12; anti-IL-12 antibodies
etc., rIL-12; recombinant IL-12 etc.). Bcl6 and IL-21 co-expression were measured by flow cytometry (d, n=4) and quantified (e, n=4), IL-21 production was quantified by ELISA (f, n=7).
One-way ANOVA with post hoc correction for multiple comparison. Error bars are mean ± SEM. (*; p<0.05, **; p<0.01). SUPPLEMENTARY FIGURE 3 ENDOSOMAL RECEPTORS ARE REQUIRED FOR THE
PRODUCTION OF IL-12 IN RESPONSE TO EC. (A) Human monocytes were left untreated (-) or treated with cytochalsin-D (CytD) to block actin polymerization and phagocytosis, or with bafilomycin A
(Baf) to inhibit phagolysosomal acidification. Cells were subsequently stimulated with EC or HKEC. IL-6 and IL-12p40 release was measured by ELISA (n=3). (B) Expression of genes encoding for
TLR1-9 was measured by qPCR in purified human monocytes, and expressed semi-quantitatively as Ct-ratio of house keeping gene _beta-actin_/_Tlr_ (n=2). (C) Human monocytes were left
untreated (ctrl), treated with pLA or pLA plus bacterial RNA (pLA+RNA) and upregulation of activation markers was measured by flow cytometry (n=5). Error bars are mean ± SEM. SUPPLEMENTARY
FIGURE 4 BACTERIAL RNA INDUCES BCL6/IL-21 EXPRESSION, ST AND HKST EQUALLY INDUCE TBET AND FOXP3 EXPRESSION. (A) Purified CD14+ porcine monocytes were stimlated with bacterial RNA complexed
with pLa, Pam3CSK4 (200ng/ml) or LPS (2ug/ml). Supernatatnts were collected after 24h and analyzed by ELISA (n=3, 4). Error bars, maximum and minimum (*; p<0.05, **; p<0.01, ***;
p<0.001). (B, C) Tbet/FoxP3 expression was detected by flow cytometry in CD3+CD4+ T cells or CD3+CD4+CD8+ T cells from pigs immunized subcutaneously with saline (ctrl), live attenuated
_S. enterica_ serovar Typhimurium vaccine (ST) or heat killed ST (HKST). (D, E) Quantification of (b) and (c) respectively (n=6,5,5). One-way ANOVA with post hoc correction for multiple
comparison. Error bars are mean ± SEM. Error bars are mean ± SEM. (*; p<0.05). SUPPLEMENTARY FIGURE 5 VACCINATION WITH ST INDUCES B CELL FOLLICLES WITH ACTIVE GERMINAL CENTERS. (A)
Sections of paraffin embedded spleen tissues were stained for KI67. Scale bars: upper panels; 5mm; lower panels; 500µm (B) Sections of paraffin embedded lymph nodes were stained for PAX5,
KI67 and BCL-2. Scale bar= 200µm. (C) Co-immunofluorescence staining of PAX5 (red) and KI67 (green) on spleen section of pigs vaccinated with ST, HKST, or saline injected control animals
(ctrl). Nuclei were counterstained with DAPI (blue). Control sections stained only with DAPI to control for autofluorescence are shown in the bottom row. Note the high level of
autofluorescence of the red pulp (erythrocytes), whereas no interfering autofluorescence signal was detected in the lymphoid follicles. Scale bar; 50µm. SUPPLEMENTARY FIGURE 6 STRUCTURE
PREDICTION MODELS OF TLR8-A AND TLR8-G. (A, B) Ribbon diagram of 3D structures for TLR8 Isoform B TLR8-A variant (a) and Isoform B TLR8-G truncated variant (b). (C, D) Surface charge
distribution of TLR8 Isoform B TLR8-A variant (c) and Isoform B TLR8-G truncated variant (d). Negatively and positively charged surface areas are colored red and blue. SUPPLEMENTARY FIGURE 7
ALTERED CLEFTS AND CAVITIES IN THE TLR8-G VARIANT. (A, B) CASTp analysis showing 148 functional pockets of TLR8 Isoform B TLR8-A variant (a) and 135 for Isoform B TLR8-G truncated variant
(b). Pockets are coloured differently. (C, D) Rear view of TLR8 structures. Isoform B TLR8-A variant (c) and Isoform B TLR8-G truncated variant (d) showing alteration of residues in pockets
associated with dimerization. Surface diagram showing major functional clefts and cavities in TLR8. (E,F) Isoform B TLR8-A variant (e) and Isoform B TLR8-G truncated variant (f). Colours
represent different clefts arranged according to volume from largest to smallest. The TLR8-G variant is 42.9% rigid (i.e., more flexible) compared to 58% for TLR8-A. (G) Comparison of the
volumes of clefts between the two gene variants. SUPPLEMENTARY FIGURE 8 TLR8-G DISPLAYS A SLIGHT GAIN-OF-FUNCTION PHENOTYPE. PBMC from healthy donors, screened for either TLR8-A or TLR8-G,
variant stimulated with (A) the TLR8 agonist CL075 (at the indicated concentrations), (B) _Mycobacterium tuberculosis_ RNA or LPS as a negative control for 18h. Cytokine release was analyzed
by ELISA (n=16, TLR8-A n=7, TLR8-G n=9). (C) HEK-Blue Null-I reporter cells were stably transfected with either TLR8-A or TLR8-G variants and stimulated with TLR8 agonist (CL075, R848) or
LPS as a negative control for 18h. NF-kB-activation was determined in the SEAP Reporter Gene Assay. Induction values were normalized to stimulation with saline. (n=6). Two-way ANOVA with
post hoc correction for multiple comparisons. Error bars are mean ± SEM. (**; p<0.01, ****; p<0.0001). SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Figures
1-8,Supplementary Note 1. LIFE SCIENCES REPORTING SUMMARY SUPPLEMENTARY TABLES 1-4 RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Ugolini, M., Gerhard,
J., Burkert, S. _et al._ Recognition of microbial viability via TLR8 drives TFH cell differentiation and vaccine responses. _Nat Immunol_ 19, 386–396 (2018).
https://doi.org/10.1038/s41590-018-0068-4 Download citation * Received: 19 July 2016 * Accepted: 15 February 2018 * Published: 19 March 2018 * Issue Date: April 2018 * DOI:
https://doi.org/10.1038/s41590-018-0068-4 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative