Play all audios:
ABSTRACT Super-resolution structured illumination microscopy (SIM) has become a widely used method for biological imaging. Standard reconstruction algorithms, however, are prone to generate
noise-specific artifacts that limit their applicability for lower signal-to-noise data. Here we present a physically realistic noise model that explains the structured noise artifact, which
we then use to motivate new complementary reconstruction approaches. True-Wiener-filtered SIM optimizes contrast given the available signal-to-noise ratio, and flat-noise SIM fully overcomes
the structured noise artifact while maintaining resolving power. Both methods eliminate ad hoc user-adjustable reconstruction parameters in favor of physical parameters, enhancing
objectivity. The new reconstructions point to a trade-off between contrast and a natural noise appearance. This trade-off can be partly overcome by further notch filtering but at the expense
of a decrease in signal-to-noise ratio. The benefits of the proposed approaches are demonstrated on focal adhesion and tubulin samples in two and three dimensions, and on nanofabricated
fluorescent test patterns. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your
institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal
Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices
may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support
SIMILAR CONTENT BEING VIEWED BY OTHERS APPROACHING MAXIMUM RESOLUTION IN STRUCTURED ILLUMINATION MICROSCOPY VIA ACCURATE NOISE MODELING Article Open access 31 January 2025 SUPERRESOLUTION
STRUCTURED ILLUMINATION MICROSCOPY RECONSTRUCTION ALGORITHMS: A REVIEW Article Open access 12 July 2023 HIGH-FIDELITY STRUCTURED ILLUMINATION MICROSCOPY BY POINT-SPREAD-FUNCTION ENGINEERING
Article Open access 01 April 2021 DATA AVAILABILITY Data are available at https://doi.org/10.4121/12942932. CODE AVAILABILITY MATLAB code is available at https://github.com/qnano/simnoise.
ImageJ code for 2D-SIM is available at https://github.com/fairSIM. REFERENCES * Neil, M. A. A., Juskaitis, R. & Wilson, T. Method of obtaining optical sectioning by using structured
light in a conventional microscope. _Opt. Lett._ 22, 1905–1907 (1997). Article CAS PubMed Google Scholar * Heintzmann, R. & Cremer, C. Laterally modulated excitation microscopy:
improvement of resolution by using a diffraction grating. _Proc. SPIE_ 3568, 185–196 (1999). Article Google Scholar * Gustafsson, M. G. L. Surpassing the lateral resolution limit by a
factor of two using structured illumination microscopy. _J. Microsc._ 198, 82–87 (2000). Article CAS PubMed Google Scholar * Gustafsson, M. G. L. et al. Three-dimensional resolution
doubling in wide-field fluorescence microscopy by structured illumination. _Biophys. J._ 94, 4957–4970 (2008). Article CAS PubMed PubMed Central Google Scholar * Schermelleh, L. et al.
Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. _Science_ 320, 1332–1336 (2008). Article CAS PubMed PubMed Central Google Scholar
* Heintzmann, R. & Huser, T. Super-resolution structured illumination microscopy. _Chem. Rev._ 117, 13890–13908 (2017). Article CAS PubMed Google Scholar * Kner, P., Chhun, B. B.,
Griffis, E. R., Winoto, L. & Gustafsson, M. G. L. Super-resolution video microscopy of live cells by structured illumination. _Nat. Methods_ 6, 339–342 (2009). Article CAS PubMed
PubMed Central Google Scholar * Shao, L., Kner, P., Rego, E. H. & Gustafsson, M. G. L. Super-resolution 3D microscopy of live whole cells using structured illumination. _Nat. Methods_
12, 1044–1046 (2011). Article CAS Google Scholar * Fiolka, R., Shao, L., Rego, E. H., Davidson, M. W. & Gustafsson, M. G. L. Time-lapse two-color 3D imaging of live cells with doubled
resolution using structured illumination. _Proc. Natl Acad. Sci. USA_ 109, 5311–5315 (2012). Article CAS PubMed PubMed Central Google Scholar * Heintzmann, R., Jovin, T. & Cremer,
C. Saturated patterned excitation microscopy – a concept for optical resolution improvement. _J. Opt. Soc. Am. B_ 19, 1599–1609 (2002). Article Google Scholar * Gustafsson, M. G. L.
Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. _Proc. Natl Acad. Sci. USA_ 102, 13081–13086 (2005). Article CAS
PubMed PubMed Central Google Scholar * Rego, E. H. et al. Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution.
_Proc. Natl Acad. Sci. USA_ 109, E135–E143 (2012). Article CAS PubMed Google Scholar * Li, D. et al. Extended-resolution structured illumination imaging of endocytic and cytoskeletal
dynamics. _Science_ 349, aab3500 (2015). Article PubMed PubMed Central CAS Google Scholar * Wicker, K., Mandula, O., Best, G., Fiolka, R. & Heintzmann, R. Phase optimization for
structured illumination microscopy. _Opt. Express_ 21, 2032–2049 (2013). Article PubMed Google Scholar * Křížek, P., Lukeš, T., Ovesný, M., Fliegel, K. & Hagen, G. M. SIMToolbox: a
MATLAB toolbox for structured illumination fluorescence microscopy. _Bioinformatics_ 32, 318–320 (2016). PubMed Google Scholar * Müller, M., Mönkemöller, V., Hennig, S., Hübner, W. &
Huser, T. Open-source image reconstruction of super-resolution structured illumination microscopy data in ImageJ. _Nat. Commun._ 7, 10980 (2016). Article PubMed PubMed Central CAS Google
Scholar * Ball, G. et al. SIMcheck: a toolbox for successful super-resolution SIM imaging. _Sci. Rep._ 5, 15915 (2015). Article CAS PubMed PubMed Central Google Scholar * Demmerle, J.
et al. Strategic and practical guidelines for successful structured illumination microscopy. _Nat. Protoc._ 12, 988–1010 (2017). Article CAS PubMed Google Scholar * Sahl, S. J. et al.
Comment on extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. _Science_ 352, 527 (2016). Article CAS PubMed Google Scholar * Li, D. et al.
Response to comment on ‘Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics’. _Science_ 352, 527 (2016). CAS PubMed Google Scholar * Righolt, C. H.
et al. Image filtering in structured illumination microscopy using the Lukosz Bound. _Opt. Express_ 21, 24431–24451 (2013). Article PubMed Google Scholar * Fried, D. L. Noise in
photo-emission current. _Appl. Opt._ 4, 79–80 (1965). Article Google Scholar * Hu, S. et al. Structured illumination microscopy reveals focal adhesions are composed of linear subunits.
_Cytoskeleton_ 72, 235–245 (2015). Article PubMed CAS Google Scholar * Unser, M., Trus, B. L. & Steven, A. C. A new resolution criterion based on spectral signal-to-noise ratio.
_Ultramicros_ 23, 39–52 (1987). Article CAS Google Scholar * Nieuwenhuizen, R. P. J. et al. Measuring image resolution in optical nanoscopy. _Nat. Methods_ 10, 557–562 (2013). Article
CAS PubMed PubMed Central Google Scholar * Chakrova, N., Heintzmann, R., Rieger, B. & Stallinga, S. Studying different illumination patterns for resolution improvement in
fluorescence microscopy. _Opt. Express_ 23, 31367–31383 (2015). Article PubMed Google Scholar * Heintzmann, R. Estimating missing information by maximum likelihood deconvolution. _Micron_
38, 136–144 (2007). Article PubMed Google Scholar * Perez, V., Chang, B.-J. & Stelzer, E. H. K. Optimal 2D-SIM reconstruction by two filtering steps with Richardson–Lucy
deconvolution. _Sci. Rep._ 6, 37149 (2016). Article CAS PubMed PubMed Central Google Scholar * Wang, H. et al. Deep learning enables cross-modality super-resolution in fluorescence
microscopy. _Nat. Methods_ 16, 103–110 (2019). Article CAS PubMed Google Scholar * Hoffman, D. P., Slavitt, I. & Fitzpatrick, C. A. The promise and peril of deep learning in
microscopy. _Nat. Methods_ 18, 131–132 (2021). Article CAS PubMed Google Scholar * Huang, X. et al. Fast, long-term, super-resolution imaging with Hessian structured illumination
microscopy. _Nat. Biotechnol._ 36, 451–459 (2018). Article CAS PubMed Google Scholar * Markwirth, A. et al. Video-rate multi-color structured illumination microscopy with simultaneous
real-time reconstruction. _Nat. Commun._ 10, 4315 (2019). Article PubMed PubMed Central CAS Google Scholar * Ströhl, F. & Kaminski, C. F. Speed limits of structured illumination
microscopy. _Opt. Lett._ 42, 2511–2514 (2017). Article PubMed Google Scholar * Chen, B.-C. et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal
resolution. _Science_ 346, 1257998 (2014). Article PubMed PubMed Central CAS Google Scholar * van der Horst, J., Trull, A. K. & Kalkman, J. Deep-tissue label-free quantitative
optical tomography. _Optica_ 7, 1682–1689 (2020). Article Google Scholar * Boulanger, J., Pustelnik, N., Condat, L., Sengmanivong, L. & Piolot, T. Nonsmooth convex optimization for
structured illumination microscopy image reconstruction. _Inverse Prob._ 34, 095004 (2018). Article Google Scholar * Weigert, M. et al. Content-aware image restoration: pushing the limits
of fluorescence microscopy. _Nat. Methods_ 15, 1090–1097 (2018). Article CAS PubMed Google Scholar * Jin, L. et al. Deep learning enables structured illumination microscopy with low
light levels and enhanced speed. _Nat. Commun._ 11, 1934 (2020). Article CAS PubMed PubMed Central Google Scholar * Krull, A. et al. Noise2Void—learning denoising from single noisy
images. in _Proc. 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)_ 2124–2132 (IEEE, 2019). * Reinhard, M. et al. An alpha-actinin binding site of zyxin is
essential for subcellular zyxin localization and alpha-actinin recruitment. _J. Biol. Chem._ 274, 13410–13418 (1999). Article CAS PubMed Google Scholar * Suresh Babu, S. et al. Mechanism
of stretch-induced activation of the mechanotransducer zyxin in vascular cells. _Sci. Signal._ 5, ra91 (2012). Article PubMed CAS Google Scholar * Yoshigi, M., Hoffman, L. M., Jensen,
C. C., Yost, H. J. & Beckerle, M. C. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. _J. Cell Biol._ 171, 209–215
(2005). Article CAS PubMed PubMed Central Google Scholar * Schlapak, R. et al. Painting with biomolecules at the nanoscale: biofunctionalization with tunable surface densities. _Nano
Lett._ 12, 1983–1989 (2012). Article CAS PubMed Google Scholar * Enguita-Marruedo, A. et al. Live cell analyses of synaptonemal complex dynamics and chromosome movements in cultured
mouse testis tubules and embryonic ovaries. _Chromosoma_ 127, 341–359 (2018). Article CAS PubMed PubMed Central Google Scholar * Peters, A. H., Plug, A. W., van Vugt, M. J. & de
Boer, P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. _Chromosome Res._ 5, 66–68 (1997). Article CAS PubMed Google Scholar *
Schücker, K., Holm, T., Franke, C., Sauer, M. & Benavente, R. Elucidation of synaptonemal complex organization by super-resolution imaging with isotropic resolution. _Proc. Natl Acad.
Sci. USA_ 112, 2029–2033 (2015). Article PubMed PubMed Central CAS Google Scholar * Heintzmann, R. et al. Calibrating photon counts from a single image. Preprint at
https://arxiv.org/abs/1611.056541611.05654 (2016). * Bakx, J. L. Efficient computation of optical disk readout by use of the chirp z transform. _Appl. Opt._ 41, 4897–4903 (2002). Article
PubMed Google Scholar * Wicker, K. Non-iterative determination of pattern phase in structured illumination microscopy using autocorrelations in Fourier space. _Opt. Express_ 21,
24692–24701 (2013). Article PubMed Google Scholar * Stallinga & Rieger, B. Accuracy of the Gaussian Point Spread Function model in 2D localization microscopy. _Opt. Express_ 18,
24461–24476 (2010). Article CAS PubMed Google Scholar * Ingaramo, M. et al. Richardson–Lucy deconvolution as a general tool for combining images with complementary strengths.
_ChemPhysChem_ 15, 794–800 (2014). Article CAS PubMed PubMed Central Google Scholar * Ströhl, F. & Kaminski, C. F. A joint Richardson–Lucy deconvolution algorithm for the
reconstruction of multifocal structured illumination microscopy data. _Methods Appl. Fluoresc._ 3, 014002 (2015). Article PubMed Google Scholar Download references ACKNOWLEDGEMENTS We
thank M. Booth and B. Rieger for stimulating research advice, A. York for suggesting the binomial random splitting of Poisson-distributed variables and W. Baarends for kindly providing
mCherry-SYCP3 samples. C.S. was supported by a Junior Research Fellowship through Merton College (Oxford, UK). L.S. acknowledges support by the Wellcome Trust Strategic Award 107457 and the
European Research Council MSC ITN grant no. 766181. N.C. acknowledges European Research Council grant no. 648580. C.H. acknowledges support from the Netherlands Organization for Scientific
Research (ZonMW-435002021). S.H. acknowledges support by NanoNextNL, a consortium of the Dutch government and 130 public and private partners. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS *
Department of Imaging Physics, Delft University of Technology, Delft, the Netherlands Carlas S. Smith, Nadya Chakrova, Sangeetha Hari, Yoram Vos, Cornelis W. Hagen, Jacob P. Hoogenboom &
Sjoerd Stallinga * Department of Physiology, Anatomy and Genetics, Centre for Neural Circuits and Behaviour, University of Oxford, Oxford, UK Carlas S. Smith * Department of Pathology,
Erasmus Optical Imaging Centre, Erasmus Medical Center, Rotterdam, the Netherlands Johan A. Slotman, Wiggert van Cappellen & Adriaan B. Houtsmuller * Micron Advanced Bioimaging Unit,
Department of Biochemistry, University of Oxford, Oxford, UK Lothar Schermelleh * Biochemistry, Molecular and Structural Biology Section, Leuven University, Leuven, Belgium Marcel Müller
Authors * Carlas S. Smith View author publications You can also search for this author inPubMed Google Scholar * Johan A. Slotman View author publications You can also search for this author
inPubMed Google Scholar * Lothar Schermelleh View author publications You can also search for this author inPubMed Google Scholar * Nadya Chakrova View author publications You can also
search for this author inPubMed Google Scholar * Sangeetha Hari View author publications You can also search for this author inPubMed Google Scholar * Yoram Vos View author publications You
can also search for this author inPubMed Google Scholar * Cornelis W. Hagen View author publications You can also search for this author inPubMed Google Scholar * Marcel Müller View author
publications You can also search for this author inPubMed Google Scholar * Wiggert van Cappellen View author publications You can also search for this author inPubMed Google Scholar *
Adriaan B. Houtsmuller View author publications You can also search for this author inPubMed Google Scholar * Jacob P. Hoogenboom View author publications You can also search for this author
inPubMed Google Scholar * Sjoerd Stallinga View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Imaging experiments were done by C.S.S.,
J.A.S., L.S., N.C., W.v.C. and A.B.H. J.A.S., S.H., Y.V., C.W.H. and J.P.H. designed and manufactured nanofabricated test samples. C.S.S., N.C., M.M. and S.S. analyzed data. S.S. derived
theory, wrote the paper and supervised the research. All authors read and approved the manuscript. CORRESPONDING AUTHOR Correspondence to Sjoerd Stallinga. ETHICS DECLARATIONS COMPETING
INTERESTS The authors declare no competing financial interests. ADDITIONAL INFORMATION PEER REVIEW INFORMATION _Nature Methods_ thanks Florian Ströhl and the other, anonymous, reviewer(s)
for their contribution to the peer review of this work. Rita Strack was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of
the editorial team. PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. EXTENDED DATA EXTENDED DATA FIG.
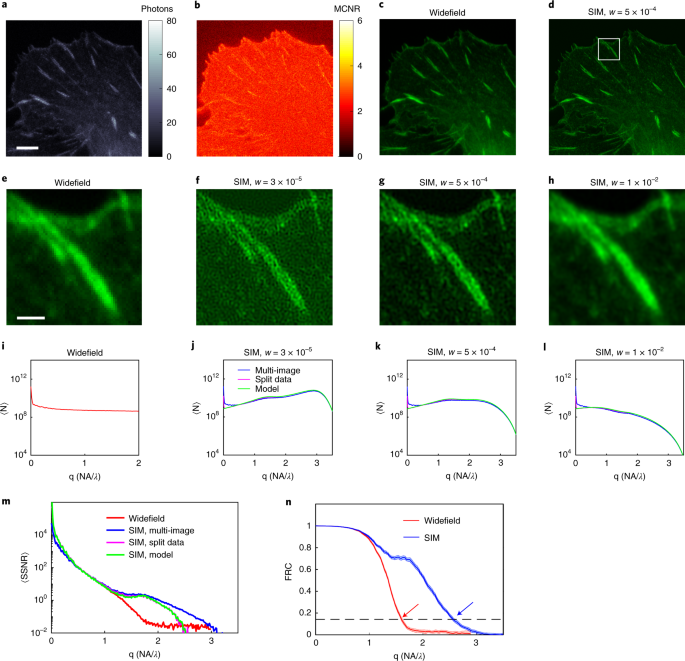
1 NOISE-CONTROLLED SIM RECONSTRUCTIONS OF GFP-ZYXIN PROTEIN IN FOCAL ADHESIONS (GREEN) AND NOISE FRACTION MAP (MAGENTA) OVER FULL FOV. A–D, State-of-art SIM (_w_ = 5 × 10−4), true-Wiener
SIM, flat-noise SIM, and notch-filtered SIM reconstructions. Contours of the noise fraction map are added in white with contour level indicated. In all reconstructions the noise fraction is
lowest in the foreground features and highest in the background region outside the cell. Overall, flat-noise SIM and true-Wiener SIM offer the lowest, and notch-filtered SIM the highest
noise enhancement. Scale bar 5 μm. EXTENDED DATA FIG. 2 MULTI-COLOR NOISE-CONTROLLED 2D-SIM RECONSTRUCTIONS. A, Combined widefield, true-Wiener SIM, flat-noise SIM, and notch-filtered SIM
reconstructions of a fluorescent test slide of a bovine pulmonary artery endothelial cell (red channel: mitochondria labeled with MitoTracker Red, green channel: actin labeled with Alexa
Fluor 488, blue channel: DNA labeled with DAPI). Note that due to embedding in hardening mounting medium, cells are flattened and 3D nuclear morphology is compromised. B-F, Insets of the
DAPI channel comparing state-of-the-art SIM with clear noise amplification artifact to the noise-controlled SIM reconstructions. The SSNR in the DAPI channel is low in this example case, due
to reduced signal intensity and compromised morphology. The low SSNR is properly taken into account by the noise-controlled SIM reconstructions, without introducing artifacts, but not by
the state-of-the-art SIM reconstruction. Scale bar (A) 10 μm, scale bar (B-F) 5 μm. EXTENDED DATA FIG. 3 NOISE PROPAGATION IN DMD-SIM. A, Reconstructions of Alexa Fluor 488 labeled actin
filaments in a bovine pulmonary artery endothelial cell with the iterative pattern-illuminated Fourier Ptychography (piFP) algorithm (see Supplementary Note 1) and with a band-pass
regularization approach for flat-noise SIM. B, Comparison of flat-noise SIM to a widefield reconstruction obtained by summing the whole set of acquired images. C-E, Insets of the boxed
region in (A) and (B). Both piFP and flat-noise SIM offer a resolution improvement, but piFP has better contrast than flat-noise SIM. The piFP reconstruction shows corrugated line structures
and punctuated features (upper right of insets), similar to the structured noise artifact in state-of-the-art SIM with line illumination patterns, flat-noise SIM shows this to a lesser
degree. Scale bar (A,B) 10 μm, scale bar (C-E) 4 μm. EXTENDED DATA FIG. 4 FLAT-NOISE SIM PROVIDES BETTER VISIBILITY OF HIGH SPATIAL FREQUENCY STRUCTURES. A–D, Widefield, true-Wiener,
flat-noise, and notch-filtered SIM reconstructions of a nanofabricated test structure of lines with 140 nm pitch. The line pattern is just visible in flat-noise, and notch-filtered SIM but
overshadowed by the noise pattern with uneven distribution of noise over spatial frequencies in true-Wiener SIM. Scale bar 1 μm. EXTENDED DATA FIG. 5 NOISE-CONTROLLED 2D-SIM OF SYNAPTONEMAL
COMPLEX. A–D, Widefield, and true-Wiener, flat-noise and notch-filtered SIM reconstructions of the mCherry-CSYCP3 protein in the synaptonemal complex. E–H, Line profiles along the lines
indicated in (B). The SIM reconstructions reveal the two cable sub-structure with a line distance of around 200 nm, flat-noise SIM has less contrast but shows smoother lines and no
background noise structure. Scale bar 3 μm. EXTENDED DATA FIG. 6 CROSS-SECTIONS IN XY AND YZ-PLANES OF 3D-RECONSTRUCTIONS OF TUBULIN (GREEN) AND NOISE FRACTION MAPS (MAGENTA) FOR DIFFERENT
CAMERA EXPOSURE TIMES. A, Widefield, (B–D) state-of-the-art SIM for low, medium and high regularization, (E) true-Wiener SIM, (F) flat-noise SIM, and (G) notch-filtered SIM. The dashed lines
in (A) indicate the location of the xz and xy slices. Scale bar 3 μm. EXTENDED DATA FIG. 7 WIDEFIELD AND NOISE-CONTROLLED 3D-SIM RECONSTRUCTIONS OF A 100 NM BEAD LAYER SAMPLE. A,B,
Widefield, (C,D) true-Wiener SIM, (E,F) flat-noise SIM, (G,H) notch-filtered SIM. The white box in (C) indicates the insets (B,D,F,H). I,J, _SSNR_ of the SIM reconstructions without (I) and
with (J) notch filtering. The data is averaged over rings in Fourier space and the plot is on a logarithmic scale according to log10(1+_SSNR_). The red line indicates the (ring averaged)
support of the SIM-OTF, the white line indicates the _SSNR_ = 5 region in Fourier space used for the extrapolation of the true-Wiener regularization filter. K, FRC curves for SIM obtained
from 4 repeated acquisitions of the bead layer sample. The FRC resolution is 106.3 ± 0.5 nm, very close to the extended SIM diffraction limit 1/(2_NA_/_λ_ + 2/_p_) = 99 nm for the estimated
pattern pitch _p_ = 416 nm, consistent with the relatively high signal level (peak pixel intensities above 104 detected photons) and the broad support of _SSNR_ above one in spatial
frequency space. Scale bar (A,C,E,G) 3 μm, scale bar (B,D,F,H) 1 μm. EXTENDED DATA FIG. 8 WIDEFIELD AND 3D NOISE-CONTROLLED SIM RECONSTRUCTIONS OF A BOVINE PULMONARY ARTERY ENDOTHELIAL CELL.
A, Widefield, (B) true-Wiener SIM, (C) flat-noise SIM, (D) notch-filtered SIM (BPAEC, red channel: mitochondria labeled with Alexa Fluor 594, green channel: actin labeled with FITC, blue
channel: DNA labeled with DAPI). Scale bar 5 μm. EXTENDED DATA FIG. 9 WIDEFIELD AND 3D NOISE-CONTROLLED SIM RECONSTRUCTIONS OF A MOUSE C127 CELL. A, Widefield, (B) true-Wiener SIM, (C)
flat-noise SIM, (D) notch-filtered SIM (magenta channel: DNA labeled with DAPI, green channel: H3K4me3 labeled with Alexa Fluor 488, blue channel: DNA labeled with DAPI). Scale bar 5 μm.
EXTENDED DATA FIG. 10 Widefield, state-of-the-art SIM and noise-controlled 3D-SIM reconstructions for one timeframe of the 15 timeframe, 7-layer dataset of H2B-GFP histone in a live HeLa
cell. Scale bar 6 μm. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Figs. 1–7 and Note. REPORTING SUMMARY SUPPLEMENTARY VIDEO 1 Impact of regularization parameter on
state-of-the-art SIM. SUPPLEMENTARY VIDEO 2 Noise-controlled SIM of 10× repeated acquisition of GFP-zyxin. SUPPLEMENTARY VIDEO 3 Noise controlled 3D SIM of tubulin, signal level 1.
SUPPLEMENTARY VIDEO 4 Noise controlled 3D SIM of tubulin, signal level 2. SUPPLEMENTARY VIDEO 5 Noise controlled 3D SIM of tubulin, signal level 3. SUPPLEMENTARY VIDEO6 Noise controlled
3D-SIM of tubulin, signal level 4. SUPPLEMENTARY VIDEO 7 Noise controlled 3D three-color SIM of BPAEC cell. SUPPLEMENTARY VIDEO 8 Noise controlled 3D two-color SIM of C127 cell.
SUPPLEMENTARY VIDEO 9 Noise controlled 3D-SIM of live HeLa cell. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Smith, C.S., Slotman, J.A., Schermelleh,
L. _et al._ Structured illumination microscopy with noise-controlled image reconstructions. _Nat Methods_ 18, 821–828 (2021). https://doi.org/10.1038/s41592-021-01167-7 Download citation *
Received: 04 September 2020 * Accepted: 26 April 2021 * Published: 14 June 2021 * Issue Date: July 2021 * DOI: https://doi.org/10.1038/s41592-021-01167-7 SHARE THIS ARTICLE Anyone you share
the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer
Nature SharedIt content-sharing initiative