Play all audios:
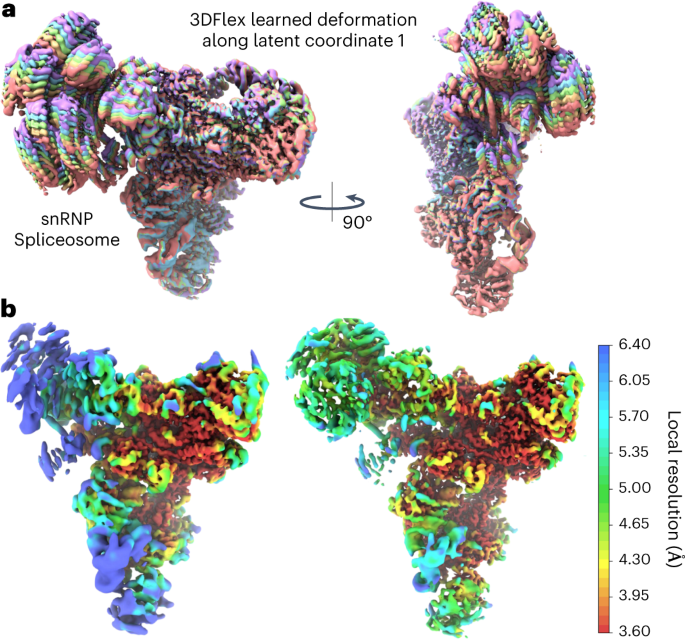
A deep learning algorithm maps out the continuous conformational changes of flexible protein molecules from single-particle cryo-electron microscopy images, allowing the visualization of the
conformational landscape of a protein with improved resolution of its moving parts. Access through your institution Buy or subscribe This is a preview of subscription content, access via
your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $32.99 / 30 days
cancel any time Learn more Subscribe to this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink *
Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional
subscriptions * Read our FAQs * Contact customer support REFERENCES * Lyumkis, D. Challenges and opportunities in cryo-EM single-particle analysis. _Biol. Chem._ 29, 5181–5197 (2019). THIS
REVIEW ARTICLE PROVIDES AN OVERVIEW OF SINGLE-PARTICLE CRYO-EM FUNDAMENTALS AND THE IMPORTANCE OF MAPPING OUT CONFORMATIONAL HETEROGENEITY. Article Google Scholar * Punjani, A. &
Fleet, D. J. 3D Variability analysis: directly resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM images. _J. Struct. Biol._ 213, 107702 (2021). THIS
PAPER REPORTS A COMPUTATIONAL METHOD FOR FITTING A LINEAR SUBSPACE MODEL OF CRYO-EM DENSITY TO SINGLE PARTICLE DATA AND IS AN IMPORTANT BASELINE FOR METHODS THAT RESOLVE CONTINUOUS
HETEROGENEITY. Article CAS PubMed Google Scholar * Zhong, E. D., Bepler, T., Berger, B. & Davis, J. H. CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural
networks. _Nat. Methods_ 18, 176–185 (2021). THIS ARTICLE REPORTS A DEEP GENERATIVE MODEL OF CRYO-EM DENSITY MAPS THAT CAN EFFECTIVELY MAP OUT NON-LINEAR CHANGES ACROSS A CONFORMATIONAL
LANDSCAPE. Article CAS PubMed PubMed Central Google Scholar * Chen, M. & Ludtke, S. J. Deep learning-based mixed-dimensional Gaussian mixture model for characterizing variability in
cryo-EM. _Nat. Methods_ 18, 930–936 (2021). THIS PAPER REPORTS A DEEP GENERATIVE MODEL THAT CAN REPRESENT THE MOTION OF PARTICLE DENSITY USING THE DISPLACEMENT OF GAUSSIAN MIXTURE
COMPONENTS. Article CAS PubMed PubMed Central Google Scholar * Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. CryoSPARC: Algorithms for rapid unsupervised cryo-EM
structure determination. _Nat. Methods_ 14, 290–296 (2017). THIS ARTICLE REPORTS THE DEVELOPMENT OF THE CRYOSPARC SOFTWARE SYSTEM, IN WHICH 3DFLEX IS IMPLEMENTED. Article CAS PubMed
Google Scholar Download references ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional
affiliations. THIS IS A SUMMARY OF: Punjani, A. & Fleet, D. J. 3DFlex: determining structure and motion of flexible proteins from cryo-EM. _Nat. Methods_
https://doi.org/10.1038/s41592-023-01853-8 (2023). RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Mapping the motion and structure of flexible proteins
from cryo-EM data. _Nat Methods_ 20, 797–798 (2023). https://doi.org/10.1038/s41592-023-01883-2 Download citation * Published: 12 May 2023 * Issue Date: June 2023 * DOI:
https://doi.org/10.1038/s41592-023-01883-2 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative