Play all audios:
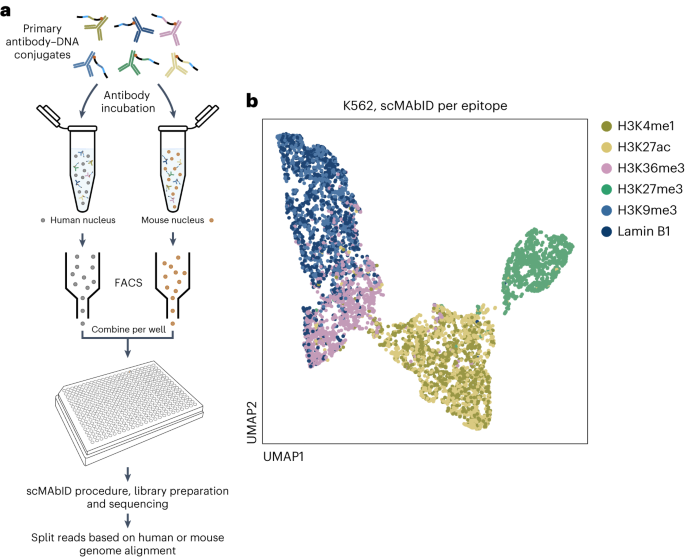
We developed MAbID, a method for combined genomic profiling of histone modifications and chromatin-binding proteins in single cells, enabling researchers to study the interconnectivity
between gene-regulatory mechanisms. We demonstrated MAbID’s implementation in profiling multifactorial changes in chromatin signatures during in vitro neural differentiation and in primary
mouse bone marrow tissue. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your
institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to this journal
Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices
may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support
REFERENCES * Vandereyken, K. et al. Methods and applications for single-cell and spatial multi-omics. _Nat. Rev. Genet._ 24, 494–515 (2023). A REVIEW ARTICLE PROVIDING AN OVERVIEW OF
MULTI-OMIC AND SINGLE-CELL SEQUENCING TECHNOLOGIES. Article CAS PubMed Google Scholar * Gopalan, S. et al. Simultaneous profiling of multiple chromatin proteins in the same cells. _Mol.
Cell_ 81, 4736–4746 (2021). ONE OF THE FIRST APPROACHES TO MEASURE SEVERAL HISTONE MODIFICATIONS IN THE SAME CELL BY IMPLEMENTING TN5-TAGMENTATION. Article CAS PubMed PubMed Central
Google Scholar * Meers, M. P. et al. Multifactorial profiling of epigenetic landscapes at single-cell resolution using MulTI-Tag. _Nat. Biotechnol._ 41, 708–716 (2023). A MULTIFACTORIAL
METHOD USING ANTIBODY–ADAPTER CONJUGATES IN COMBINATION WITH TN5. Article CAS PubMed Google Scholar * Bartosovic, M. & Castelo-Branco, G. Multimodal chromatin profiling using
nanobody-based single-cell CUT&Tag. _Nat. Biotechnol._ 41, 794–805 (2023). A METHOD COMBINING MULTIFACTORIAL PROFILING WITH ATAC-SEQ USING NANOBODY–TN5 FUSIONS. Article CAS PubMed
Google Scholar * Handa, T. et al. Chromatin integration labeling for mapping DNA-binding proteins and modifications with low input. _Nat. Protoc._ 15, 3334–3360 (2020). THIS PROTOCOL
DESCRIBES THE GENERATION OF UNIQUELY BARCODED ANTIBODY–DNA CONJUGATES. Article CAS PubMed Google Scholar Download references ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature
remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. THIS IS A SUMMARY OF: Lochs, S. J. A. et al. Combinatorial single-cell profiling of
major chromatin types with MAbID. _Nat. Methods_ https://doi.org/10.1038/s41592-023-02090-9 (2023). RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE
Multifactorial epigenomic profiling of six chromatin states in single cells. _Nat Methods_ 21, 20–21 (2024). https://doi.org/10.1038/s41592-023-02091-8 Download citation * Published: 04
December 2023 * Issue Date: January 2024 * DOI: https://doi.org/10.1038/s41592-023-02091-8 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get
shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative