Play all audios:
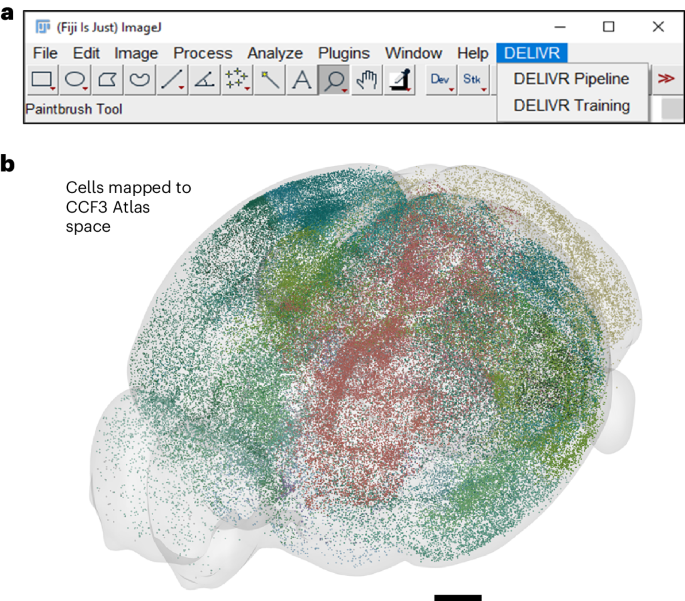
We created DELiVR, a deep-learning pipeline for 3D brain-cell mapping that is trained with virtual reality-generated reference annotations. It can be deployed via the user-friendly interface
of the open-source software Fiji, which makes the analysis of large-scale 3D brain images widely accessible to scientists without computational expertise. Access through your institution
Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 print issues and online access $259.00 per year only
$21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout
ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support REFERENCES * Molbay, M. et al. A guidebook for DISCO tissue
clearing. _Mol. Syst. Biol._ 17, e9807 (2021). A REVIEW THAT PRESENTS AN OVERVIEW OF TISSUE CLEARING AND LSFM. Article PubMed PubMed Central Google Scholar * Renier, N. et al. Mapping of
brain activity by automated volume analysis of immediate early genes. _Cell_ 165, 1789–1802 (2016). THIS PAPER PRESENTS CLEARMAP, A THRESHOLD-BASED METHOD FOR BRAIN ACTIVITY MAPPING.
Article CAS PubMed PubMed Central Google Scholar * Al-Maskari, R. et al. On the pitfalls of deep image segmentation for lightsheet microscopy. In _Medical Imaging with Deep Learning_
https://openreview.net/forum?id=3Krfu84W-Wx (2022). THIS SHORT REVIEW SUMMARIZES CHALLENGES FOR SEGMENTING STRUCTURES IMAGED THROUGH LSFM. * Pidhorskyi, S. et al. syGlass: Interactive
exploration of multidimensional images using virtual reality head-mounted displays. Preprint at https://doi.org/10.48550/arXiv.1804.08197 (2018). THIS PAPER DESCRIBES THE DEVELOPMENT OF A
SOFTWARE PACKAGE FOR VISUALIZING VOLUMETRIC DATA WITH VR HEADSETS. * Todorov, M. I. et al. Machine learning analysis of whole mouse brain vasculature. _Nat. Methods._ 17, 442–449 (2020).
THIS PAPER PRESENTS A MACHINE LEARNING-BASED APPROACH FOR THE SEGMENTATION OF THE ENTIRE VASCULATURE IN WHOLE MOUSE BRAINS. Article CAS PubMed PubMed Central Google Scholar Download
references ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. THIS IS A SUMMARY
OF: Kaltenecker, D. et al. Virtual reality-empowered deep-learning analysis of brain cells. _Nat_. _Methods_ https://doi.org/10.1038/s41592-024-02245-2 (2024). RIGHTS AND PERMISSIONS
Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Simplifying deep learning to enhance accessibility of large-scale 3D brain imaging analysis. _Nat Methods_ 21, 1151–1152 (2024).
https://doi.org/10.1038/s41592-024-02246-1 Download citation * Published: 22 April 2024 * Issue Date: July 2024 * DOI: https://doi.org/10.1038/s41592-024-02246-1 SHARE THIS ARTICLE Anyone
you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the
Springer Nature SharedIt content-sharing initiative