Play all audios:
ABSTRACT Metagenomic experiments expose the wide range of microscopic organisms in any microbial environment through high-throughput DNA sequencing. The computational analysis of the
sequencing data is critical for the accurate and complete characterization of the microbial community. To facilitate efficient and reproducible metagenomic analysis, we introduce a
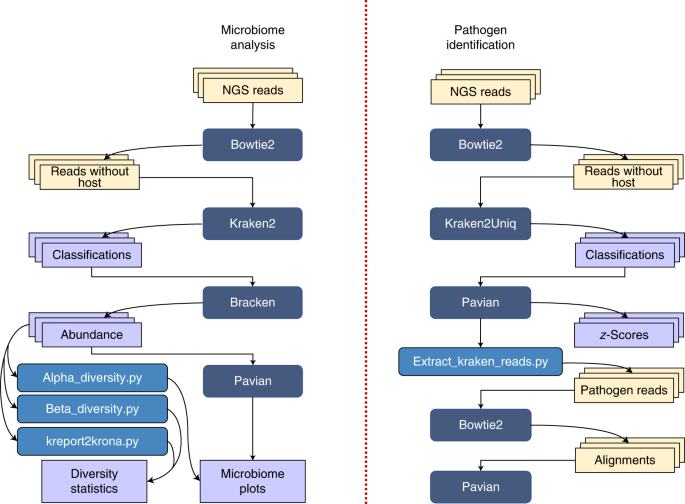
step-by-step protocol for the Kraken suite, an end-to-end pipeline for the classification, quantification and visualization of metagenomic datasets. Our protocol describes the execution of
the Kraken programs, via a sequence of easy-to-use scripts, in two scenarios: (1) quantification of the species in a given metagenomics sample; and (2) detection of a pathogenic agent from a
clinical sample taken from a human patient. The protocol, which is executed within 1–2 h, is targeted to biologists and clinicians working in microbiome or metagenomics analysis who are
familiar with the Unix command-line environment. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access
through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to
this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy
now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact
customer support SIMILAR CONTENT BEING VIEWED BY OTHERS BENCHMARKING SECOND AND THIRD-GENERATION SEQUENCING PLATFORMS FOR MICROBIAL METAGENOMICS Article Open access 11 November 2022 CRITICAL
ASSESSMENT OF METAGENOME INTERPRETATION: THE SECOND ROUND OF CHALLENGES Article Open access 08 April 2022 UNVEILING MICROBIAL DIVERSITY: HARNESSING LONG-READ SEQUENCING TECHNOLOGY Article
30 April 2024 DATA AVAILABILITY The microbiome analysis used three samples from Taur et al.8, and the pathogen identification used ten samples from Li et al.9, all of which can be found on
NCBI with their SRA IDs. Source data are provided with this paper. CODE AVAILABILITY The following website details and links all software and databases used in this protocol:
http://ccb.jhu.edu/data/kraken2_protocol/. We also provide easy-to-use Jupyter notebooks for both workflows, which can be executed in the browser using Google Collab:
https://github.com/martin-steinegger/kraken-protocol/. CHANGE HISTORY * _ 29 AUGUST 2024 A Correction to this paper has been published: https://doi.org/10.1038/s41596-024-01064-1 _
REFERENCES * Rappé, M. S. & Giovannoni, S. J.The uncultured microbial majority. _Annu. Rev. Microbiol._ 57, 369–394 (2003). Article PubMed Google Scholar * Wood, D. E. & Salzberg,
S. L.Kraken: ultrafast metagenomic sequence classification using exact alignments. _Genome Biol._ 15, R46 (2014). Article PubMed PubMed Central Google Scholar * Breitwieser, F. P.,
Baker, D. N. & Salzberg, S. L.KrakenUniq: confident and fast metagenomics classification using unique k-mer counts. _Genome Biol._ 19, 198 (2018). Article CAS PubMed PubMed Central
Google Scholar * Wood, D. E., Lu, J. & Langmead, B. Improved metagenomic analysis with Kraken 2. _Genome Biol._ 20, 257 (2019). Article CAS PubMed PubMed Central Google Scholar *
Lu, J., Breitwieser, F. P., Thielen, P. & Salzberg, S. L.Bracken: estimating species abundance in metagenomics data. _PeerJ Comput. Sci._ 3, e104 (2017). Article Google Scholar *
Breitwieser, P. & Salzberg, S. L.Pavian: interactive analysis of metagenomics data for microbiome studies and pathogen identification. _Bioinformatics_ 36, 1303–1304 (2020). Article CAS
PubMed Google Scholar * Langmead, B. & Salzberg, S. L.Fast gapped-read alignment with Bowtie 2. _Nat. Methods_ 9, 357–359 (2012). Article CAS PubMed PubMed Central Google Scholar
* Taur, Y. et al.Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. _Sci. Transl. Med._ 10, eaap9489 (2018). Article PubMed
PubMed Central Google Scholar * Li, Z. et al.Identifying corneal infections in formalin-fixed specimens using next generation sequencing. _Invest. Ophthalmol. Vis. Sci._ 59(Jan), 280–288
(2018). Article CAS PubMed PubMed Central Google Scholar * Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J.Basic local alignment search tool. _J. Mol. Biol._
215(Oct), 403–410 (1990). Article CAS PubMed Google Scholar * Pruitt, K. D., Tatusova, T. & Maglott, D. R.NCBI reference sequences (RefSeq): a curated non-redundant sequence database
of genomes, transcripts and proteins. _Nucleic Acids Res._ 35, D61–D65 (2007). Article CAS PubMed Google Scholar * O’Leary, N. A. et al.Reference sequence (RefSeq) database at NCBI:
current status, taxonomic expansion, and functional annotation. _Nucleic Acids Res._ 44, D733–D745 (2016). Article PubMed Google Scholar * Ounit, R., Wanamaker, S., Close, T. J. &
Lonardi, S.CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative _k_-mers. _BMC Genomics_ 16, 236 (2015). Article PubMed PubMed Central Google
Scholar * Kim, D., Song, L., Breitwieser, F. P. & Salzberg, S. L.Centrifuge: rapid and sensitive classification of metagenomic sequences. _Genome Res._ 26, 1721–1729 (2016). Article
CAS PubMed PubMed Central Google Scholar * Menzel, P., Ng, K. L. & Krogh, A.Fast and sensitive taxonomic classification for metagenomics with Kaiju. _Nat. Commun._ 7, 11257 (2016).
Article CAS PubMed PubMed Central Google Scholar * Ye, S. H., Siddle, K. J., Park, D. J. & Sabeti, P. C.Benchmarking metagenomics tools for taxonomic classification. _Cell_ 178,
779–794 (2019). Article CAS PubMed PubMed Central Google Scholar * Seppey, M., Manni, M. & Zdobnov, M.LEMMI: a continuous benchmarking platform for metagenomics classifiers. _Genome
Res._ 30, 1208–1216 (2020). Article CAS PubMed PubMed Central Google Scholar * Segata, N. et al.Metagenomic microbial community profiling using unique clade-specific marker genes.
_Nat. Methods_ 9, 811–814 (2012). Article CAS PubMed PubMed Central Google Scholar * Vervier, K., Mahé, P., Tournoud, M., Veyrieras, J. B. & Vert, J. P.Large-scale machine learning
for metagenomics sequence classification. _Bioinformatics_ 32, 1023–1032 (2016). Article CAS PubMed Google Scholar * Luo, Y., Yu, Y. W., Zeng, J., Berger, B. & Peng, J.Metagenomic
binning through low-density hashing. _Bioinformatics_ 35, 219–226 (2019). Article CAS PubMed Google Scholar * Breitwieser, F. P., Lu, J. & Salzberg, S. L.A review of methods and
databases for metagenomic classification and assembly. _Brief. Bioinform._ 20, 1125–1136 (2017). Article PubMed Central Google Scholar * Li, H. Aligning sequence reads, clone sequences
and assembly contigs with BWA-MEM. Preprint at _arXiv_ https://doi.org/10.48550/arXiv.1303.3997 (2013). * Li, H.Minimap2: pairwise alignment for nucleotide sequences. _Bioinformatics_ 34,
3094–3100 (2018). Article CAS PubMed PubMed Central Google Scholar * Stephens, Z. et al.Exogene: a performant workflow for detecting viral integrations from paired-end next-generation
sequencing data. _PLoS ONE_ 16, e0250915 (2021). Article CAS PubMed PubMed Central Google Scholar * Breitwieser, F. P., Pertea, M., Zimin, A. V. & Salzberg, S. L.Human contamination
in bacterial genomes has created thousands of spurious proteins. _Genome Res._ 29, 954–960 (2019). Article CAS PubMed PubMed Central Google Scholar * Steinegger, M. & Salzberg, S.
L.Terminating contamination: large-scale search identifies more than 2,000,000 contaminated entries in GenBank. _Genome Biol._ 21, 115 (2020). Article CAS PubMed PubMed Central Google
Scholar * Lu, J. & Salzberg, S. L.Removing contaminants from databases of draft genomes. _PLoS Comput. Biol._ 14, e1006277 (2018). Article PubMed PubMed Central Google Scholar *
Buchfink, B., Xie, C. & Huson, D. H.Fast and sensitive protein alignment using DIAMOND. _Nat. Methods_ 12, 59–60 (2015). Article CAS PubMed Google Scholar * Mirdita, M., Steinegger,
M., Breitwieser, F., Söding, J. & Levy Karin, E. Fast and sensitive taxonomic assignment to metagenomic contigs. _Bioinformatics_ 37, 3029–3031 (2021). Article CAS PubMed PubMed
Central Google Scholar * Nasko, D. J., Koren, S., Phillippy, A. M. & Treangen, T. J.RefSeq database growth influences the accuracy of _k_-mer-based lowest common ancestor species
identification. _Genome Biol._ 19, 165 (2018). Article PubMed PubMed Central Google Scholar * Yang, C. et al.A review of computational tools for generating metagenome-assembled genomes
from metagenomic sequencing data. _Comput. Struct. Biotechnol. J._ 19, 6301–6314 (2021). Article CAS PubMed PubMed Central Google Scholar * Whittaker, R. H.Evolution and measurement of
species diversity. _Taxon_ 21, 213–251 (1972). Article Google Scholar * Berger, W. H. & Parker, F. L. Diversity of planktonic foraminifera in deep-sea sediments. _Science_ 168,
1345–1347 (1970). Article CAS PubMed Google Scholar * Fisher, R. A., Corbet, A. S. & Williams, C. B.The relation between the number of species and the number of individuals in a
random sample of an animal population. _J. Anim. Ecol._ 12, 42–58 (1943). Article Google Scholar * Simpson, E. H.Measurement of diversity. _Nature_ 163, 688–688 (1949). Article Google
Scholar * Shannon, C. E.A mathematical theory of communication. _Bell Syst. Tech. J._ 27, 379–423 (1948). Article Google Scholar * Bray, J. R. & Curtis, J. T.An ordination of the
upland forest communities of southern Wisconsin. _Ecol. Monogr._ 27, 325–349 (1957). Article Google Scholar * Ondov, B. D., Bergman, N. H. & Phillippy, A. M.Interactive metagenomic
visualization in a web browser. _BMC Bioinform._ 12, 385 (2011). Article Google Scholar * Danecek, P. et al.Twelve years of SAMtools and BCFtools. _Gigascience_ 10, giab008 (2021). Article
PubMed PubMed Central Google Scholar * Grüning, B. et al.Bioconda: sustainable and comprehensive software distribution for the life sciences. _Nat. Methods_ 15, 475–476 (2018). Article
PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS Indexes for tools in the Kraken suite, including the indexes used in this protocol, are made freely available
on Amazon Web Services thanks to the AWS Public Dataset Program. B.L. was supported by NIH/NIHMS grant R35GM139602. S.L.S. was supported by NIH grants R35-GM130151 and R01-HG006677. M.S.
acknowledges support from the National Research Foundation of Korea grant (2019R1A6A1A10073437, 2020M3A9G7103933, 2021R1C1C102065 and 2021M3A9I4021220); New Faculty Startup Fund; and the
Creative-Pioneering Researchers Program through Seoul National University. AUTHOR INFORMATION Author notes * These authors contributed equally: Jennifer Lu, Natalia Rincon. AUTHORS AND
AFFILIATIONS * Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, USA Jennifer Lu, Natalia Rincon & Steven L. Salzberg * Center for Computational Biology,
Whiting School of Engineering, Johns Hopkins University, Baltimore, MD, USA Jennifer Lu, Natalia Rincon, Derrick E. Wood, Florian P. Breitwieser, Christopher Pockrandt & Steven L.
Salzberg * Department of Computer Science, Johns Hopkins University, Baltimore, MD, USA Derrick E. Wood, Ben Langmead & Steven L. Salzberg * Department of Biostatistics, Johns Hopkins
University, Baltimore, MD, USA Steven L. Salzberg * School of Biological Sciences and Institute of Molecular Biology & Genetics, Seoul National University, Seoul, Republic of Korea
Martin Steinegger Authors * Jennifer Lu View author publications You can also search for this author inPubMed Google Scholar * Natalia Rincon View author publications You can also search for
this author inPubMed Google Scholar * Derrick E. Wood View author publications You can also search for this author inPubMed Google Scholar * Florian P. Breitwieser View author publications
You can also search for this author inPubMed Google Scholar * Christopher Pockrandt View author publications You can also search for this author inPubMed Google Scholar * Ben Langmead View
author publications You can also search for this author inPubMed Google Scholar * Steven L. Salzberg View author publications You can also search for this author inPubMed Google Scholar *
Martin Steinegger View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS J.L. and M.S. led the development of the protocol. N.R. executed and
designed the microbiome analysis protocol and is the author of the KrakenTools α-diversity tools. J.L. developed the pathogen identification protocol and is the author of Bracken and
KrakenTools. M.S. authored the Jupyter notebooks for the protocol. D.E.W. is the senior author of Kraken and Kraken 2. F.B. is the author of KrakenUniq. C.P. is an author for the KrakenTools
β-diversity script. B.L. supervised the development of Kraken 2. S.L.S. supervised the development of Kraken, KrakenUniq and Bracken. B.L. and S.L.S. supervised the development of this
protocol. All authors contributed to the writing of the manuscript. CORRESPONDING AUTHORS Correspondence to Jennifer Lu or Martin Steinegger. ETHICS DECLARATIONS COMPETING INTERESTS The
authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Protocols_ thanks the anonymous reviewers for their contribution to the peer review of this work.
ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. RELATED LINKS KEY REFERENCES
USING THIS PROTOCOL Salzberg, S. et al. _Neurol. Neuroimmunol. Neuroinflamm_. 3, e251 (2016): https://doi.org/10.1212/NXI.0000000000000251 Wood, D. et al. _Genome Biol_. 15, R46 (2014):
https://doi.org/10.1186/gb-2014-15-3-r46 Lu, J. et al. _Peer J. Comput. Sci_. 3, e104 (2017): https://doi.org/10.7717/peerj-cs.104 Breitwieser, F. et al. _Genome Biol_. 19, 198 (2018):
https://doi.org/10.1186/s13059-018-1568-0 Wood, D. et al. _Genome Biol_. 20, 257 (2019): https://doi.org/10.1186/s13059-019-1891-0 Breitwieser, F. et al. _Bioinformatics_ 36, 1303–1304
(2020): https://doi.org/10.1093/bioinformatics/btz715 KEY DATA USED IN THIS PROTOCOL Taur, Y. et al. _Sci. Transl. Med_. 10, eaap9489 (2018): https://doi.org/10.1126/scitranslmed.aap9489 Li,
Z. et al. _Invest_. _Ophthalmol. Vis. Sci_. 59, 280–288 (2018): https://doi.org/10.1167/iovs.17-21617 SUPPLEMENTARY INFORMATION SUPPLEMENTARY TABLE 1 Supplementary Table 1 SUPPLEMENTARY
TABLE 2 Supplementary Table 2 SOURCE DATA SOURCE DATA FIG. 2 Breport text for plotting Sankey, and krona counts for plotting krona plots. SOURCE DATA FIG. 6 Alpha diversity table text, bray
Curtis equation text, and heatmap values for beta diversity. SOURCE DATA FIG. 7 Pathogen sample species heat map data. RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a society
or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of
this article is solely governed by the terms of such publishing agreement and applicable law. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Lu, J., Rincon, N., Wood, D.E.
_et al._ Metagenome analysis using the Kraken software suite. _Nat Protoc_ 17, 2815–2839 (2022). https://doi.org/10.1038/s41596-022-00738-y Download citation * Received: 29 June 2021 *
Accepted: 16 June 2022 * Published: 28 September 2022 * Issue Date: December 2022 * DOI: https://doi.org/10.1038/s41596-022-00738-y SHARE THIS ARTICLE Anyone you share the following link
with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt
content-sharing initiative