Play all audios:
Correction to _Scientific Reports_ https://doi.org/10.1038/s41598-020-64664-3, published online 12 May 2020 The original version of this Article contained errors in the qPCR analysis of the
gene expression levels and mitochondrial DNA content. The calculations were based on the incorrect assumption that the amplification efficiency was 100%. The Authors have now reanalysed the
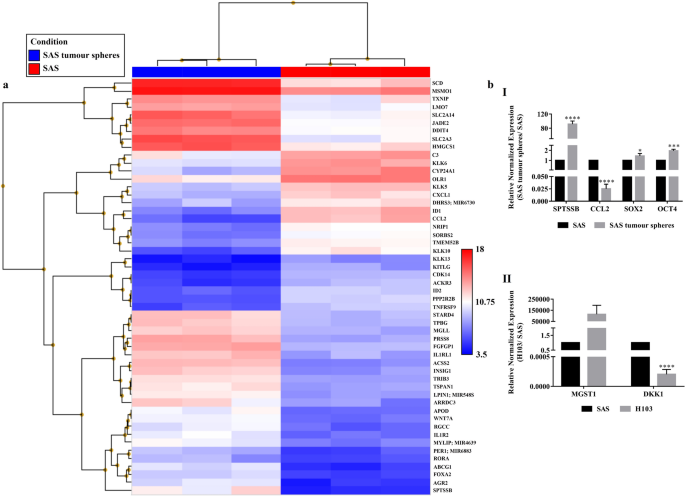
data using the actual values for amplification efficiency (estimated using the serial dilution method), and found that the p-values marked by asterisks in panel (b) of Figure 2 and Figure 3
and described in the figure legends were incorrect. The description of the statistical significance annotations in these figures has been corrected in the figure legends, where in the legend
of Figure 2, “Data are presented as mean ± SD. *P < 0.05, ***P < 0.001, ****P < 0.0001, n = 3.” now reads: “Data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P <
0.001, n = 3.” In the legend of Figure 3, “Data are presented as mean ± SD. **P < 0.01, ***P < 0.001, n = 3.” now reads: “Data are presented as mean ± SD. **P < 0.01, n = 3.”
Figures 2 and 3 and their accompanying legends have been corrected in the published Article, the original versions appear below for reference. Additionally, in the Results and discussion
section under the subheading ‘SAS tumour spheres with stem cell-like features showed increased expressions of metabolism-associated and pluripotency genes.’, “The increased expression levels
of two of the genes, namely _OCT4_ (P = 0.03) and _SOX2_ (P = 0.0004), and the proteins they encode (Oct4, P = 0.004; Sox2, P = 0.17) were confirmed by qPCR (Fig. 2b) and Western blotting
(Fig. 1c).” now reads: “The increased expression levels of two of the genes, namely _OCT4_ (P = 0.016) and _SOX2_ (P = 0.052), and the proteins they encode (Oct4, P = 0.004; Sox2, P = 0.17)
were confirmed by qPCR (Fig. 2b) and Western blotting (Fig. 1c).” Furthermore, the Data availability statement was incomplete, where “Some of the raw data has been provided as supplementary
datasets.” now reads: “The raw sequencing data generated by MinKNOW were deposited in Sequence Read Archive (SRA; Accession No.: PRJNA712949). The raw data from the microarray analysis were
deposited in Gene Expression Omnibus (GEO; Accession No.: GSE168424). The raw data for the other analyses are provided as supplementary datasets.” Finally, Supplementary Tables S6 and S7 and
Supplementary Datasets S4 and S6 have been corrected to include the values for amplification efficiency. The original Supplementary Information file which includes Tables S6 and S7, and the
original Supplementary Datasets S4 and S6 are provided below. The original Article has been corrected. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Drug and Herbal Research Centre, Faculty
of Pharmacy, Universiti Kebangsaan Malaysia, Jalan Raja Muda Abdul Aziz, 50300, Kuala Lumpur, Malaysia Amnani Aminuddin, Pei Yuen Ng & Eng Wee Chua * School of Pharmacy, International
Medical University, Bukit Jalil, 57000, Kuala Lumpur, Malaysia Chee-Onn Leong * Centre for Cancer and Stem Cell Research, Institute for Research, Development and Innovation, International
Medical University, Bukit Jalil, 57000, Kuala Lumpur, Malaysia Chee-Onn Leong Authors * Amnani Aminuddin View author publications You can also search for this author inPubMed Google Scholar
* Pei Yuen Ng View author publications You can also search for this author inPubMed Google Scholar * Chee-Onn Leong View author publications You can also search for this author inPubMed
Google Scholar * Eng Wee Chua View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Eng Wee Chua. SUPPLEMENTARY
INFORMATION SUPPLEMENTARY INFORMATION. DATASET S4. DATASET S6. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License,
which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link
to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence,
unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory
regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Aminuddin, A., Ng, P.Y., Leong, CO. _et al._ Author Correction: Mitochondrial DNA
alterations may influence the cisplatin responsiveness of oral squamous cell carcinoma. _Sci Rep_ 11, 14193 (2021). https://doi.org/10.1038/s41598-021-93444-w Download citation * Published:
05 July 2021 * DOI: https://doi.org/10.1038/s41598-021-93444-w SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a
shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative