Play all audios:
ABSTRACT Black _Aspergillus_ species are the most common etiological agents of otomycosis, and pulmonary aspergillosis. However, limited data is available on their antifungal susceptibility
profiles and associated resistance mechanisms. Here, we determined the azole susceptibility profiles of black _Aspergillus_ species isolated from the Indian environment and explored the
potential resistance mechanisms through _cyp51A_ gene sequencing, protein homology modeling, and expression analysis of selected genes _cyp51A_, _cyp51B_, _mdr1_, and _mfs_ based on their
role in imparting resistance against antifungal drugs_._ In this study, we have isolated a total of 161 black aspergilli isolates from 174 agricultural soil samples. Isolates had variable
resistance towards medical azoles; approximately 11.80%, 3.10%, and 1.24% of isolates were resistant to itraconazole (ITC), posaconazole (POS), and voriconazole (VRC), respectively. Further,
_cyp51A_ sequence analysis showed that non-synonymous mutations were present in 20 azole-resistant _Aspergillus_ section _Nigri_ and 10 susceptible isolates. However, Cyp51A homology
modeling indicated insignificant protein structural variations because of these mutations. Most of the isolates showed the overexpression of _mdr1_, and _mfs_ genes. Hence, the study
concluded that azole-resistance in section _Nigri_ cannot be attributed exclusively to the _cyp51A_ gene mutation or its overexpression. However, overexpression of _mdr1_ and _mfs_ genes may
have a potential role in drug resistance. SIMILAR CONTENT BEING VIEWED BY OTHERS EPIDEMIOLOGY, ANTIFUNGAL SUSCEPTIBILITY AND BIOLOGICAL CHARACTERISTICS OF CLINICAL _ASPERGILLUS FUMIGATUS_
IN A TERTIARY HOSPITAL Article Open access 15 May 2025 MECHANISMS OF MULTIDRUG RESISTANCE CAUSED BY AN IPI1 MUTATION IN THE FUNGAL PATHOGEN _CANDIDA GLABRATA_ Article Open access 25 January
2025 _FKS1_ MUTATION ASSOCIATED WITH DECREASED ECHINOCANDIN SUSCEPTIBILITY OF _ASPERGILLUS FUMIGATUS_ FOLLOWING ANIDULAFUNGIN EXPOSURE Article Open access 20 July 2020 INTRODUCTION
_Aspergillus niger_ and its related species are grouped in _Aspergillus_ section _Nigri_, commonly known as black aspergilli. These species are unevenly distributed globally and are often
isolated from clinical samples1. Black aspergilli infections are the third most common cause of _Aspergillus_-associated infections_,_ leading to conditions such as otomycosis,
onychomycosis, and pulmonary aspergillosis2,3. Triazoles comprise the first-line treatment for aspergillosis however, long-term therapy and widespread use of azole-based pesticides in
agriculture have raised concerns because of an increase in resistance to the medical triazoles in various _Aspergillus_ species4. The acquired resistance is not only because of the
fungicidal effect of azoles but also caused by azole exposure in clinical and environmental settings5,6,7,8. Multi-azole resistance has been reported in patients and from the environment
across Europe, China Japan, The Middle East, and India5,9,10,11,12. Several authors have reported azole-resistant _Aspergillus_ isolates, which correlates with the poor therapeutic outcome
of azole, thereby limiting the treatment options6,7,13,14. Previous studies have reported that the antifungal drug susceptibility of itraconazole (ITC) against clinical and environmental
isolates of _A. niger_ and _A. tubingensis_ showed higher minimum inhibitory concentration (MIC)15,16,17,18. The azole drugs act via non-competitive binding to the Cyp51 enzyme, a sterol
14α-demethylase, of the ergosterol biosynthetic pathway. The azole inhibits ergosterol synthesis and disrupts cell membrane19. The mechanism of azole resistance in _Aspergillus fumigatus_
has been extensively studied_._ Several mutations in the _cyp51A_ gene and overexpression of this gene have been reported in azole-resistant _A. fumigatus_ strains20. However, azole
resistance mechanisms have not been extensively investigated in the _A. niger_ complex. The molecular mechanism of azole resistance in section _Nigri_ was first reported by Howard et al.15.
Since then, a few reports have elaborated on the resistance mechanisms against _A. niger_ complex via mutation analysis of the _cyp51A_ gene or its expression. Pérez-Cantero et al.21
reported that _cyp51B_ gene expression in the _Aspergillus_ section _Nigri_ was not inducible after azole exposure. However, the underlying azole resistance mechanisms in _Aspergillus_
section _Nigri_ have not been fully explored. Identification of a relatively large number of clinical azole-resistant _A. fumigatus_ isolates lacking the _cyp51A_ mutations and comparative
genomics studies in yeasts prompted the investigations on alternative mechanisms of azole resistance. This led to the discovery of the role of efflux pumps in azole resistance. Efflux pumps
are categorized into two main classes: the major-facilitator superfamily (MFS) proteins, encoded by 278 genes, and ATP-binding cassette (ABC) proteins, encoded by 49 genes22. Tobin et al.23
identified two ABC transporter proteins; MDR1 and MDR2 in _A. fumigatus_ based on cloning and sequence homology. Further, overexpression of MDR3 and MDR4 in ITC-resistant _A. fumigatus_
strains was reported24,25. In a recent study, overexpression of efflux pump genes has been reported in azole resistant _A. niger_ isolates26. Overexpression of efflux pump genes has also
been observed in response to amphotericin B in _A. fumigatus._ This report suggested that the fungus adapts by overexpressing genes and proteins involved in drug efflux, representing a
mechanism to develop resistance and survive against antifungal drugs27. Moreover, overexpression of efflux pumps genes like _abcC_ in azole-resistant _A. fumigatus_ isolates has been
highlighted in other studies28,29. Here, we aimed to determine the correlation between azole resistance and the _cyp51A_ gene mutation in black _Aspergillus_ isolates isolated from
agricultural soil samples across India through antifungal susceptibility testing, gene sequencing, expression analysis of selected genes and protein homology modeling. Further, we attempted
to elucidate the role of efflux transporter genes, _mdr1_, and _mfs_, in azole-resistant environmental isolates. RESULTS IDENTIFICATION AND ANTIFUNGAL SUSCEPTIBILITY TESTING We screened 161
black aspergilli isolates isolated from 174 soil samples across India followed by antifungal susceptibility analysis against ITC, voriconazole (VRC), and posaconazole (POS). Results revealed
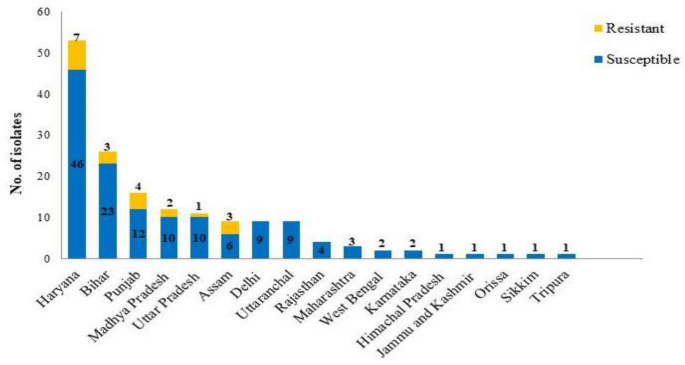
that out of 161 isolates, 20 were resistance to at least one azole drug. Resistant isolates were obtained from the samples collected from Haryana (7/46), Bihar (3/23), Punjab (4/12), Madhya
Pradesh (2/10), Uttar Pradesh (1/10 and Assam (3/6) region (Fig. 1). Furthermore, azole susceptibility revealed that 19/161 black aspergilli isolates was above its epidemiological cutoff
value (ECV) of > 2 μg/mL for ITC. Five isolates had MIC > 0.5 μg/mL for POS (Table 1; Fig. 2). Four black aspergilli isolates were cross-resistant to ITC and POS. For VRC, all isolates
(except AG1 and AU3) showed MICs below the ECV values (2 μg/mL), and only two isolates were cross-resistant to ITC and VRC (AU3 and AG1). We inferred that VRC and POS were the most
effective triazoles against black aspergilli isolates, and ITC was the least effective triazole. MOLECULAR IDENTIFICATION OF BLACK _ASPERGILLUS_ ISOLATES Molecular identification was
performed for 20 azole-resistant black aspergilli isolates. Additionally, to provide a comparative context, we included 10 azole-susceptible black aspergilli isolates from the same
geographical region as the resistant isolates. Using 18S internal transcribed spacer (ITS) and _β_-tubulin gene sequencing a total of 21 _A. niger_ (13 resistant and 8 susceptible) and 9 _A.
tubingensis_ (7 resistant and 2 susceptible) isolates from the 30 _Aspergillus_ section _Nigri_ isolates were identified. All tested ITS and _β_-tubulin gene sequences displayed 99% to 100%
nucleotide identity with the sequences available in NCBI database. The GenBank accession numbers generated for the submitted ITS sequences were OK342205, MZ305300, MZ305301, MZ305302
MZ305303, MZ292199, MZ292200, MZ292198, MZ292197, MZ292196, MW332573, MW282896, OQ938544, OQ938545, OQ938546, OQ935460, OQ935461, OQ935462, OQ935463, OQ935464, OQ935465, OQ935466, OQ935467,
OR575635, OR575636, OR575637, OR575638, OR575639, OR575640 and for _β_-tubulin gene were OQ948174, OQ948175, OQ948176, OQ948177, OQ948178, OQ948179, OQ948180, OQ948181, OQ948182, OQ948183,
OQ948184, OQ948185, OQ948186, OQ948187, OQ948188, OQ948189, OQ948189, OQ948190, OQ948191, OQ948192, OQ948193, OQ948194,OQ948195, OR584016, OR584017, OR584018, OR584019, OR584020, OR584021,
OR584022, OR584023. MUTATION ANALYSIS OF THE _CYP51A_ GENE The _cyp51A_ gene was sequenced and analyzed for all 20 resistant (13 _A. niger_ and 7 _A. tubingensis_) and 10 susceptible (8 _A.
niger_ and 2 _A. tubingensis_) _Aspergillus_ section _Nigri_ isolates. The sequences of the _cyp51A_ gene of _A. niger_ and _A. tubingensis_ resistant isolates were aligned with the
wild-type strains NT_166526 and JF450924.1, respectively (Supplementary Figs. S1, S2). We identified 15 non-synonymous mutations among the _Aspergillus_ section _Nigri_ isolates. Table 2
shows the amino acid alterations in the _cyp51A_ gene. Among these, several mutations were found in both susceptible and resistant isolates. In _A. niger_, the amino acid change Q228R was
observed in both resistant and susceptible isolates. However, the mutation S346R in combination with Q228R was identified in various isolates, with different susceptibility to azole drugs.
This included one azole-susceptible isolate (PU2), two isolates resistant to either ITC or POS (RK5 and MP6), two isolates displaying cross-resistance to ITC and VRC (AU3 and AG1), and two
isolates with cross resistance to ITC and POS (AU4 and MP8). Further, the amino acid change V383L, R501Q, I244F, E254D, D253Y, T267Y, Y268H, E278Q and L303M were found only in three _A.
niger_ resistant isolates, that exhibited resistance to only ITC (MD2, PR4, PI5).In the case of _A. tubingensis,_ amino acid substitution T321A in combination with V377I was observed in
three ITC resistant isolates (FA9, FA4, and FA6). Additionally, one of the identified ITC resistant isolate (FA10) exhibited this substitution along with K477N mutation. Mutation T321A was
also observed independently in ITC-resistant isolate (AH6), an isolate exhibiting cross resistant to both ITC and POS (PR2), and a susceptible isolate (AH1). Furthermore, a new amino acid
substitution, V329I was detected in one ITC-resistant (PR9) and in one susceptible (UT1) _A. tubingensis_ isolates. CYP51A HOMOLOGY MODELING AND MOLECULAR DOCKING We constructed ten
_Aspergillus_ section _Nigri_ Cyp51A homology models (A to L) to compare the amino acid profiles of azole-resistant isolates with those of wild-type isolates. Five mutation combinations
observed in all _A. niger_ isolates were: Q228R/V383L, Q228R/S346R, Q228R, Q228R/R501Q, and Q228R/I244F/D253Y/E254D/T267Y/Y268H/E278Q. Therefore, models A to E were constructed after
incorporating these mutations in _A. niger_ isolates (Fig. 3). Similarly, _A. tubingensis_ models G to K were constructed for the mutations V329I/L492M, T321A, V377I/T321A, V329I, and
K477N/T321A (Fig. 3), and two wild-type models F and L was constructed for _A. niger_ and _A. tubingensis_, respectively. Table 3 showed the docking score of the different Cyp51A protein
models of the aspergilli isolates with ITC and VRC. Compared to wild-type isolates, marked variations in the overall protein structure conferring resistance were not identified in models A
to E, and G to K. The most negative docking score was obtained with model E for ITC, and docking scores were almost similar in all the models for VRC. H-bond interactions between the ligands
(ITC and VRC) and Cyp51A protein structure were obtained in models A, D, H, J, K, and L. In model A, ITC interacted with CYS447, in model H, it interacted with TYR51, and in model K
interaction was observed between ITC and TYR119, whereas, two H-bonds were formed between model J and ITC with TYR119 and SER360. VRC interacted with TYR119 via H-bond interactions in models
A, H, I, J, K, and L, while, H-bond interactions were observed between CYS447 and VRC in models D and G. The obtained homology-modeled protein of each isolate was aligned individually with
wild-type Cyp51A modeled protein. The differences in protein backbone structures are quantitated with the root mean square deviation (RMSD) of the similarity between two superimposed atomic
coordinates of modelled proteins. The RMSD scores demonstrated high similarity among models A to E when compared to the susceptible _A. niger_ model (model F), with all models exhibiting a
difference of less than 0.1 Å. In the case of _A. tubingensis_, the RMSD scores also demonstrated substantial similarity among models G to K compared to the susceptible _A. tubingensis_
model (model L), all models displaying a difference of less than 0.24 Å (Fig. 3). Table 4 depicted the RMSD values obtained (because of mutations) of the aligned protein models of the
mutated sequences with wild-type Cyp51A of _A. niger_ and _A. tubingensis_, respectively. GENE EXPRESSION ANALYSIS Reverse transcription analysis from the RNA samples followed by qRT-PCR was
performed to understand the expression modulation of transcripts encoding for _cyp51A_, _cyp51B, mdr1_, and _mfs_ genes in the azole-resistant isolates of _Aspergillus_ section _Nigri_.
Figure 4 indicates the two-fold relative expression of crucial genes in _Aspergillus_ section _Nigri_ isolates compared to the susceptible isolates PI2 of _A. niger_ and UT2 of _A.
tubingensis._ Results depicted a consistent upregulation (> 3 folds) of _cyp51A_ gene in isolates exhibiting exclusive resistance to ITC (MD2, PU6, PI5, and PR4), with the exception of
PR3 and F1 isolates. Additionally, a slight upregulation (1.5 folds) of this gene was observed in a ITC resistant isolate RK5. Upregulation was also noticed in two out of the three isolates
(AU4 and PI3) that showed cross-resistance to ITC and POS. Notably, the _cyp51A_ gene showed a 1.7 folds upregulation in a susceptible isolate (PR5). However, in isolates with
cross-resistance to ITC and VRC or isolate resistant only to POS, no upregulation was observed. In case of _A. tubingensis_ isolates, the _cyp51A_ gene consistently showed upregulation in
all the isolates, except the susceptible isolate AH1. The expression analysis of the _cyp51B_ gene revealed downregulation in all _A. niger_ isolates including susceptible and resistant
isolates while the gene was > 2 folds upregulated in three azole-resistant (FA4, FA9 and FA10) _A. tubingensis_ isolates_._ Relative expression of gene _mdr1_ showed over 2 folds
increased expression in two ITC resistant isolates (PU6, and PI5) and one ITC resistant isolate (RK5) with a 1.5 folds upregulation. Additionally, there was a 4.19 folds upregulation in an
isolate displaying cross-resistance to ITC and VRC (AU3), and 2 folds upregulation in an isolate with cross-resistance to ITC and POS (AU4). In the case of _A. tubingensis,_ the gene was 2
folds upregulated only in two ITC resistant isolates (FA4 and FA9).Among the 13 _A. niger_ isolates resistant to azoles, the expression of the _mfs_ gene was found to be upregulated in five
isolates resistant to ITC, three isolates displaying cross-resistance to ITC and POS, in one isolate resistant to POS, and one isolate demonstrating cross-resistance to ITC and VRC. Notably,
the _mfs_ gene exhibited high expression, with a 10 folds increase in isolate MD2, 6.19 folds in isolate F1, and a 5 folds upregulation in MP6 and MP8. Whereas, the _mfs_ gene was
downregulated in all _A. tubingensis_ isolates_._ It’s noteworthy that overexpression of both the _mdr1_ and _mfs_ gene was not observed in any of the susceptible _Aspergillus_ section
_Nigri_ isolates. The expression data were normalized by housekeeping gene actin. DISCUSSION The members of _Aspergillus_ section _Nigri_ causes several human diseases such as keratitis and
invasive aspergillosis30,31. However, the in vivo efficacy of antifungal therapy against the clinical isolates of black aspergilli is undetermined, and _in-vitro_ data is limited. Several
authors from different countries have highlighted the prevalence of azole resistance in _A. fumigatus_4,32,33. However, limited number of studies has reported _in-vitro_ antifungal
susceptibilities and resistance mechanisms in the black _Aspergillus_ spp. In our study, we isolated 161 black aspergilli from soil samples across diverse regions in India. Azole
susceptibility assay identified 20 azole resistant isolates. Notably, the number of the resistant isolates was prominent in states known for extensive agricultural activities, including
Haryana (7/46), Bihar (3/23), Punjab (4/12), Madhya Pradesh (2/10), Uttar Pradesh (1/10), and Assam (3/6). We hypothesized that this prevalence might be linked to the use of azole fungicides
in these regions. This hypothesis aligns with similar observations in studies involving azole-resistant _A. fumigatus_ isolates found in soils exposed to fungicides34,35. The absence of
resistant isolates in other states like West Bengal despite being a major agriculture state may be due to the limited number of samples collected. Further studies with more extensive
sampling are warranted. Azole susceptibility testing of these 161 isolates demonstrated that ITC resistance (19/161) was more common suggesting that ITC resistance may be more obvious among
the species of black aspergilli. Similar result was observed in previous study conducted by Howard et al.15. In contrast, POS was the most effective azole against _Aspergillus_ section
_Nigri_ isolates. Several authors have reported similar results17,32,36. Interestingly, cross-resistance for the three tested azole drugs were not obtained in our study. Further, the
molecular identification of these 30 isolates revealed that 21 as _A. niger_ and 9 isolates as _A. tubingensis_. Other _Aspergillus_ section _Nigri_ isolates, such as _A. welwitschiae_ and
_A. brasiliensis_, were not identified in our dataset, despite being present in previous studies21,37. This discrepancy may be attributed to variations in geographical distribution. We also
tried to correlate the relation between azole drug resistance and mutations in the _cyp51A_ gene of _Aspergillus_ section _Nigri_ isolates in this study. Sequence analysis of the _cyp51A_
gene revealed that amino acid alterations, including Q228R, S346R, V329I, and T321A were present in susceptible and resistant _Aspergillus_ section _Nigri_ isolates. Several new mutations
(R501Q, I244F, E254D, D253Y, T267Y, Y268H, E278Q, and L303M) have been observed in ITC resistant isolates only. However, it remains unclear whether these mutations are responsible for azole
resistance in these isolates or not, Hence, Cyp51A protein modeling was conducted to further examine the role of these mutations in azole resistance. Molecular docking results revealed
docking scores for the drugs, and the most negative docking score was obtained with model E (Mutations, Q228R/I244F/D253Y/E254/D/T267Y/Y268H/E278Q) for ITC. The docking score was almost
similar in all the models for VRC. Docking results suggested no significant change in the binding site of the Cyp51A protein due to amino acid substitutions. The modeling results and
similarity between the protein structures (RMSD score) suggested that _cyp51A_ mutations in these isolates do not cause a marked change in the overall protein structure and do not directly
interfere with their binding to azole drugs to confer resistance. Despite the _in-silico_ findings, it is crucial to acknowledge the limitations of computational models. To validate the real
impact of these amino acid substitutions, further _in-vitro_ experiments employing the CRISPR/Cas9 system are essential. The approach has previously been employed to study the _cyp51A_ gene
in _Aspergillus_ species38,39. This study also demonstrated that mutations Q228R, S346R, and T321A were not specific to a particular azole drug. The presence of these mutations were
observed in different isolates displaying varied resistance viz_,_ ITC or POS, as well as in isolates exhibiting cross-resistance to ITC and VRC or ITC and POS. This diversity in mutation
patterns suggested a lack of uniformity in the correlation between specific mutations and resistance profiles. Similar to the study conducted by Howard et al.15, which also proposed that
mutations in the _cyp51A_ gene may not be crucial for azole resistance in section Nigri. Azole resistance is mainly associated with acquiring genetic mutations or overexpression of the
_cyp51A_ gene and genes associated with efflux pump40. The overexpression of the _cyp51A_ gene plays a crucial role in azole resistance in _A. fumigatus_; therefore, the expression of the
_cyp51A_ gene was also investigated in all 30 _Aspergillus_ section _Nigri_ isolates. Our findings revealed that the expression of _cyp51A_ gene was significantly (_p_ value ≤ 0.05)
upregulated in seven azole-resistant _A. niger_ isolates when compared to the susceptible isolate PI2. Similarly, the gene exhibited overexpression in all the resistant _A. tubingensis_
isolates in comparison to the susceptible isolate UT2. Surprisingly, even a susceptible _A. niger_ isolate (PR5) carrying the Q228R mutation in the _cyp51A_ displayed significant
overexpression of this gene. This observation implies that the overexpression of the _cyp51A_ gene may not always result in azole resistance in this fungal species. These results were
consistent with the observations reported in previous studies21,37. Furthermore, we found that the _cyp51B_ gene was downregulated in both the resistant and susceptible _A. niger_ isolates
in comparison to susceptible isolate PI2. Conversely, in three ITC-resistant isolates (FA4, FA9, and FA10), characterized by the presence of T321A in combination with V377I or K477N
mutations, where the _cyp51B_ gene exhibited upregulation compared to susceptible UT2 isolate. The baseline expression of _cyp51A_ was more than that of _cyp51B._ However, a recent study on
ergosterol quantification has revealed that both enzymes have a comparable impact on the total ergosterol content within the _Aspergillus_ section _Nigri_ cell41. In _Candida albicans_ and
_Candida glabrata_, overexpression of efflux pumps, ATP-binding cassette transporters, and transporters of the major facilitator superfamily has been extensively studied42. Other studies
have highlighted the significance of efflux pump genes overexpression in azole resistant _A. fumigatus_ as well28,29. However, to the best of our knowledge, the expression of efflux pump
genes in _Aspergillus_ section _Nigri_ isolates in India has not been investigated. In a recent study overexpression of these genes in _Aspergillus_ section _Nigri_ isolates has been
reported, suggesting a possible drug resistance mechanism26. Therefore, we analyzed the expression of efflux pumps genes. The relative expression of the MDR efflux pump gene _mdr1_ was
upregulated in five _A. niger_ resistant isolates (AU3, AU4, PU6, RK5, and PI5) and two _A. tubingensis_ resistant isolates (FA4 and FA9). The overexpression of _mdr1_ gene was observed
along with the overexpression of _cyp51A_ and _mfs_ gene in 3 isolates (AU4, PU6, and RK5) carrying Q228R mutation alone or in combination with S346R. 2 folds upregulation of the gene was
also observed in PI5 isolate carrying mutation R501Q in combination with Q228R. Conversely in isolate AU3, carrying similar mutations (Q228R, S346R) showed 4 folds overexpression only for
_mdr1_ gene. Additionally, two _A. tubingensis_ resistant isolates displayed elevated expression of the _mdr1_ gene, along with overexpression of _cyp51A_ and _cyp51B_. The diverse gene
expression pattern even among isolate with similar mutation highlights the complex azole resistance mechanisms in these fungal isolates. The _mfs_ gene, was upregulated in 10 out of 13
resistant _A. niger_ isolates, and the gene was downregulated in all _A. tubingensis_ isolates_._ Remarkably, among these three _A. niger_ isolates (AG1, MP6, and MP8) displaying diverse
susceptibility patterns, a common mutation profile of Q228R in combination with S346R was associated solely with the upregulation of the _mfs_ gene in these isolates. The _mfs_ gene
exhibited high expression, with a 10 folds increase in isolate MD2, 6.19 folds in isolate F1, and a 5 folds upregulation in MP6 and MP8. However, it’s important to note that efflux pumps
genes _mdr1_ and _mfs_ was not upregulated in all the susceptible _Aspergillus_ section _Nigri_ isolates. These findings suggest a possible association between the overexpression of these
genes and drug resistance in _Aspergillus_ section _Nigri_ isolates. Previous studies have also investigated the overexpression of the _mdr1_ gene in azole-resistant _Aspergillus flavus_
isolates lacking mutations in the _cyp51A_ region43. Additionally, reports in _A. fumigatus_ have indicated the upregulation of the _mfs_ gene without _cyp51A_ mutation44. Furthermore, the
overexpression of _mdr1_ and _mfs_ genes has also been reported in other studies in _A. flavus_45,46. Although we identified mutations in the _cyp51A_ gene of _Aspergillus_ section _Nigri_
isolates but our analysis using Cyp51A homology modeling did not show significant changes in protein structure due to these mutations. Therefore, we analyzed the role of efflux pumps in
conferring resistance and found that 92% of _A. niger_ resistant isolates exhibited overexpression of efflux pumps genes either _mdr1_ or _mfs._ The overexpression of these genes may cause
azole resistance in _Aspergillus_ section _Nigri_ isolates. However, in the context of _A. tubingensis_ isolates, we noted overexpression of the efflux pump gene (_mdr1_) in only two of the
isolates. Interestingly, one of the _A. niger_ isolate (PR3) exhibited resistance to the drug ITC. However, the gene expression data for selected genes (_cyp51A, cyp51B, mdr1, and mfs_) did
not show significant overexpression in comparison to sensitive isolate. This observation suggests the presence of an alternative mechanisms contributing to azole resistance. The potential
mechanisms may involve the upregulation of ABC transporter genes such as _cdr1B_ and _mdr4,_ which could result in reduced drug concentrations with in these fungal isolates. To
comprehensively understand the reasons behind this susceptibility profile and to explore potential resistance mechanisms further experiments will be required. The excessive use of triazoles
in agriculture leads to their accumulation in the environment leading to the development of azole resistance. These resistant strains may infect the immunocompromised individuals and are
subsequently detected in clinical settings. Our previous research, as well as other studies, have reported instances of azole resistance in _A. fumigatus_ in the environment due to the use
of azole fungicides33,47. In the present study, the identification of azole-resistant _Aspergillus_ section _Nigri_ isolates from the same geographical area and the overexpression of efflux
pump genes in these isolates without exposure to azole drugs, has prompted us to hypothesize. We suggests that the emergence of resistance in _Aspergillus_ section _Nigri_ might be linked to
the environmental application of azole fungicides, potentially triggering a stress response leading to upregulation of efflux pump genes. Therefore, large-scale epidemiological studies are
required to monitor the resistant fungal strains in the environment. Further, the antifungal susceptibility profiles of environmental or clinical fungal isolates should be accurately
monitored. CONCLUSION The identified 15 amino acid substitutions in the _cyp51A_ gene of _Aspergillus_ section _Nigri_ isolates in our study does not completely correlate with resistance
data, which suggests, other genes such as _mdr/ mfs_ needs to be investigated for mutational analysis. Advanced techniques such as CRISPR/Cas9 could be applied to study the genetic changes.
The results in the study suggests that overexpression of _mdr1_ and _mfs_ genes could potentially play a role in drug resistance in _Aspergillus_ section _Nigri._ Studies on expression of
the _cyp51A, cyp51B_, and multidrug efflux transporter at transcript and protein levels will help identify other genes and proteins involved in resistance. Further, genome-wide profiling may
provide a better understanding of the mechanisms of azole resistance in _Aspergillus_ section _Nigri_ isolates. METHODS ENVIRONMENTAL SAMPLING AND ISOLATION OF BLACK _ASPERGILLI_ A total of
174 agricultural soil samples were collected from 19 states across India including Assam (n = 12), Bihar (n = 21), Chhattisgarh (n = 2), Delhi (n = 12), Haryana (n = 23), Himachal Pradesh
(n = 3), Jammu and Kashmir (n = 1), Karnataka (n = 4), Maharashtra (n = 3), Manipur (n = 3), Madhya Pradesh (n = 10), Orissa (n = 4), Punjab (n = 12), Rajasthan (n = 9), Sikkim (n = 1),
Tripura (n = 1), Uttaranchal (n = 8), Uttar Pradesh (n = 31), and West Bengal (n = 14). All samples were processed and inoculated on potato dextrose agar (PDA) plates as previously described
by Sen et al.48. The plates were incubated at 28 ± 2 °C for 5 days. All black aspergilli isolates grown on PDA plates were identified based on their macroscopic and microscopic
morphologies48. The isolates were further sub-cultured on PDA and stored at 4 °C till further use. ANTIFUNGAL SUSCEPTIBILITY TESTING The spores (conidia) of all the black aspergilli isolates
were harvested in sterile phosphate buffered saline (1 × PBS) supplemented with 0.05% Tween 20; suspension was then adjusted to 1 × 104 conidia/mL in potato dextrose broth. _In-vitro_
susceptibility testing of black aspergilli isolates was performed to determine the MICs of ITC, VRC and POS using CLSI M38- A2 broth microdilution method49 in a 96-well flat bottom
polystyrene plate (Tarsons, India). The experiment was performed in triplicate for each isolate. The azole drug stocks were prepared in dimethyl sulfoxide. Two-fold dilutions were prepared
in a 96-well microplate to obtain final concentrations ranging from 64–0.125 µg/mL for ITC, VRC, and POS. Conidial suspension (100 µL) was added to each well except negative control. The
plates were incubated statically for 4 days at 28 ± 2 °C. The proposed ECVs for ITC, VRC, and POS were 2, 2 and 0.5 µg/mL, respectively50,51. These values were used to interpret the results.
The MIC value of a drug was determined as the lowest concentration with no visible growth relative to the drug-free control. GENOMIC DNA EXTRACTION To identify the azole resistant black
_Aspergillus_ isolates and analyse their mutations related to azole susceptibility, we used the cetyl trimethyl ammonium bromide (CTAB) method52,53 to extract genomic DNA from 20 resistant
black aspergilli isolates. Additionally, to establish a comparative context, we have selected an extra set of 10 azole susceptible black _Aspergillus_ isolates from the same geographical
region as the resistant isolates. MOLECULAR IDENTIFICATION OF BLACK _ASPERGILLUS_ ISOLATES All the 30 isolates (20 resistant and 10 susceptible) were identified by the amplification and
sequencing of full-length 18S ITS region, partial sequencing of _β_-tubulin genes using the ITS1 and ITS454 and Tub5 (5' TGACCCAGCAGATGTT 3') and Tub6 (5' GTTGTTGGGAATCCACTC
3')55 primers, respectively as previously described47. SEQUENCING OF _CYP51A_ GENE Different primer sets were used for amplification of the _cyp51A_ gene of twenty resistant and ten
susceptible isolates (Table S1)21. Nucleotide sequencing was performed via Sanger’s sequencing using ABI 370 XL (Applied Biosystems). The sequence of the products was compared to the _A.
niger_ (Accession no. NT_166526) and _A. tubingensis_ (Accession no. JF450924.1) _cyp51A_ wild-type sequence using the NCBI alignment service, Align Sequence Nucleotide BLAST
(https://blast.ncbi.nlm.nih.gov/Blast.cgi) and Clustal Omega tool (https://www.ebi.ac.uk/Tools/msa/clustalo/). HOMOLOGY MODELING Cyp51A protein structure of _A. niger_ and _A. tubingensis_
were not available on RCSB-PDB hence; FASTA sequence of _A. niger_ (XP_001394224.1) and _A. tubingensis_ (AEK81606.1) were retrieved from NCBI to construct the homology models of the wild
type strains. Amino acid substitutions were incorporated in the reference protein sequences of _A. niger_ and _A. tubingensis_ to perform homology modeling. The X-ray crystal structures of
protein 4UYM and 5FRB were used as reference for the model generation of _A. niger_ and _A_. _tubingensis_ based on the sequence similarity. The Schrödinger Maestro multiple sequence viewer
(MSV) tool56 was used to construct a structure-based alignment of the mutated templates. The template structure for alignment was identified through searching the wild type sequences using
the BLAST tool incorporated in Schrodinger Maestro multiple sequence viewer. The homology models were built with Prime in Schrödinger Suite (Schrödinger, LLC, New York, NY). Models were
refined using minimization and loop refining tool of prime. The results of homology modeling were further validated using Ramachandran plot. Modeled structures were aligned and root mean
square deviation (RMSD) was calculated using Maestro Schrödinger. MOLECULAR DOCKING The 3D structures of VRC (PubChem ID- 71,616) and ITC (PubChem ID- 55283) were retrieved from the PubChem
(https://pubchem.ncbi.nlm.nih.gov/). The 3D structures of these drugs were prepared using Ligprep in Schrödinger Suite and possible states were generated at pH 7.0 ± 2.0 (LigPrep 2022). The
homology models were prepared for docking using one-step protein preparation workflow in Schrödinger Maestro (Maestro 2022) by adding and refining missing hydrogen atoms. Molecular docking
calculations were completed using Glide in Schrodinger docking suits (Glide 2021)57. Modeled proteins were prepared by restrained minimization using force field OPLS3e. Grids centers were
determined from ligands of reference proteins. Receptor grid maps representing the shape and chemical properties of the binding site were generated using Schrödinger Glide. The binding site
of modeled proteins was also confirmed by the Sitemap predicting possible binding pockets. The grid sites were created using Glide receptor grid generator with docking length of 20 Å.
Docking was performed using the Schrödinger Virtual Screening Workflow tool using Glide Standard Precision (SP) with the OPLS3e force field58. One pose per ligand was kept for post-docking
full force field minimization (optimization of ligand pose geometry followed by recalculation of interaction strength between ligand–protein using the scaled Coulomb-van der Waals term and
the Glide score). Docking scores are reported in kcal/mol, the more negative the number, the better binding. QUANTIFICATION OF GENE EXPRESSION BY QRT-PCR To assess the expression level of
the _cyp51A_, _cyp51B, mdr1,_ and _mfs_ genes in thirty _Aspergillus_ section _Nigri_ isolates, quantitative real time reverse transcription PCR (qRT-PCR) was used. Further, to carry out
qRT-PCR, RNA was extracted from the harvested mycelia using TRIzol reagent (Thermo Fisher)59,60. This experiment was carried out in the absence of azole drug exposure. The extracted RNA was
then reverse transcribed into first-stand cDNA using the Hi-cDNA synthesis kit (HiMedia, India) by following the manufacturer’s recommendations. Real-time qPCR was performed using an ABI
QuantStudio 3 (Applied Biosystems, Streetsville, Canada) as previously described in a study by Gupta et al.60. The gene expression was estimated using the 2−ΔΔCt method, with actin as the
reference gene21. Accession numbers XM_001398369.2 and XM_035497978.1 were used for the primer designing of _mdr1_ and _mfs_ gene, respectively. The gene specific primers were designed using
the Primer 3 software (http://primer3.ut.ee/). The primer sets used in this study are listed in Table S1. STATISTICAL ANALYSIS One-way ANOVA test was used to compare relative gene
expression levels. The experiment was conducted in biological and technical triplicate. The _p_ value of ≤ 0.05 was considered significant. Statistical analysis was also performed using
GraphPad Prism v8.0.2.263. DATA AVAILABILITY Data are presented within the manuscript and in the Supplementary Information. REFERENCES * Xess, I., Mohanty, S., Jain, N. & Banerjee, U.
Prevalence of Aspergillus species in clinical samples isolated in an Indian tertiary care hospital. _Indian J. Med. Sci._ 58, 513–519 (2004). PubMed Google Scholar * Pappas, P. G. _et al._
Invasive fungal infections among organ transplant recipients: results of the transplant-associated infection surveillance network (TRANSNET). _Clin. Infect. Dis._ 50, 1101–1111.
https://doi.org/10.1086/651262 (2010). Article PubMed Google Scholar * Hendrickx, M., Beguin, H. & Detandt, M. Genetic re-identification and antifungal susceptibility testing of
_Aspergillus_ section Nigri strains of the BCCM/IHEM collection. _Mycoses_ 55, 148–155 (2012). Article CAS PubMed Google Scholar * Chowdhary, A. _et al._ Multi-azole-resistant
_Aspergillus fumigatus_ in the environment in Tanzania. _J. Antimicrob. Chemother._ 69, 2979–2983. https://doi.org/10.1093/jac/dku259 (2014). Article CAS PubMed Google Scholar *
Chowdhary, A., Kathuria, S., Xu, J. & Meis, J. F. Emergence of azole-resistant _aspergillus fumigatus_ strains due to agricultural azole use creates an increasing threat to human health.
PLoS Pathog. 9, e1003633. https://doi.org/10.1371/journal.ppat.1003633. Epub 2013 Oct 24. Erratum in: PLoS Pathog. 9.
doi:https://doi.org/10.1371/annotation/4ffcf1da-b180-4149-834c-9c723c5dbf9b. (2013). * Wiederhold, N. & Patterson, T. Emergence of azole resistance in _Aspergillus_. _Semin. Respir.
Crit. Care Med._ 36, 673–680. https://doi.org/10.1055/s-0035-1562894 (2015). Article PubMed Google Scholar * Chowdhary, A., Sharma, C. & Meis, J. F. Azole-resistant aspergillosis:
Epidemiology, molecular mechanisms, and treatment. _J. Infect. Dis._ 216, S436–S444. https://doi.org/10.1093/infdis/jix210 (2017). Article CAS PubMed Google Scholar * Sen, P., Vijay, M.,
Singh, S., Hameed, S. & Vijayaraghvan, P. Understanding the environmental drivers of clinical azole resistance in _Aspergillus_ species. _Drug Target Insights_ 16, 25–35.
https://doi.org/10.33393/dti.2022.2476 (2022). Article PubMed PubMed Central Google Scholar * Lockhart, S. R. _et al._ Azole resistance in _Aspergillus fumigatus_ isolates from the
ARTEMIS global surveillance study is primarily due to the TR/L98H mutation in the cyp51A gene. _Antimicrob. Agents Chemother._ 55(9), 4465–4468. https://doi.org/10.1128/aac.00185-11 (2011).
Article CAS PubMed PubMed Central Google Scholar * Baddley, J. W. _et al._ Patterns of susceptibility of _Aspergillus_ isolates recovered from patients enrolled in the
transplant-associated infection surveillance network. _J. Clin. Microbiol._ 47(10), 3271–3275. https://doi.org/10.1128/jcm.00854-09 (2009). Article CAS PubMed PubMed Central Google
Scholar * Tashiro, M. _et al._ Antifungal susceptibilities of _Aspergillus fumigatus_ clinical isolates obtained in Nagasaki, Japan. _Antimicrob. Agents Chemother._ 56(1), 584–587.
https://doi.org/10.1128/aac.05394-11 (2012). Article CAS PubMed PubMed Central Google Scholar * Cao, D. _et al._ Five-year survey (2014 to 2018) of azole resistance in environmental
_Aspergillus fumigatus_ isolates from China. _Antimicrob. Agents Chemother._ 64, e00904-20 (2020). Article CAS PubMed PubMed Central Google Scholar * Perlin, D. S., Rautemaa-Richardson,
R. & Alastruey-Izquierdo, A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. _Lancet Infect. Dis._ 17, e383–e392.
https://doi.org/10.1016/S1473-3099(17)30316-X (2017). Article PubMed Google Scholar * Wiederhold, N. Antifungal resistance: Current trends and future strategies to combat. _Infect. Drug
Resist._ 10, 249–259. https://doi.org/10.2147/IDR.S124918 (2017). Article CAS PubMed PubMed Central Google Scholar * Howard, S. J., Harrison, E., Bowyer, P., Varga, J. & Denning, D.
W. Cryptic species and azole resistance in the _Aspergillus niger_ complex. _Antimicrob. Agents Chemother._ 55, 4802–4809. https://doi.org/10.1128/AAC.00304-11 (2011). Article CAS PubMed
PubMed Central Google Scholar * Li, Y., Wan, Z., Liu, W. & Li, R. Identification and susceptibility of _Aspergillus_ section _nigri_ in china: Prevalence of species and paradoxical
growth in response to echinocandins. _J. Clin. Microbiol._ 53, 702–705. https://doi.org/10.1128/JCM.03233-14 (2015). Article PubMed PubMed Central Google Scholar * Iatta, R. _et al._
Species distribution and in vitro azole susceptibility of _Aspergillus_ Section _Nigri_ isolates from clinical and environmental settings. _J. Clin. Microbiol._ 54, 2365–2372.
https://doi.org/10.1128/JCM.01075-16 (2016). Article CAS PubMed PubMed Central Google Scholar * Mirhendi, H., Zarei, F., Motamedi, M. & Nouripour-Sisakht, S. _Aspergillus
tubingensis_ and _Aspergillus niger_ as the dominant black _Aspergillus_, use of simple PCR-RFLP for preliminary differentiation. _J. Mycol. Med._ 26, 9–16.
https://doi.org/10.1016/j.mycmed.2015.12.004 (2016). Article CAS PubMed Google Scholar * Parker, J. E. _et al._ Resistance to antifungals that target CYP51. _J. Chem. Biol._ 7, 143–161.
https://doi.org/10.1007/s12154-014-0121-1 (2014). Article PubMed PubMed Central Google Scholar * Price, C. L., Parker, J. E., Warrilow, A. G., Kelly, D. E. & Kelly, S. L. Azole
fungicides—Understanding resistance mechanisms in agricultural fungal pathogens. _Pest Manag. Sci._ 71, 1054–1058. https://doi.org/10.1002/ps.4029 (2015). Article CAS PubMed Google
Scholar * Pérez-Cantero, A., López-Fernández, L., Guarro, J. & Capilla, J. New insights into the Cyp51 contribution to azole resistance in _Aspergillus_ section _Nigri_. _Antimicrob.
Agents Chemother._ 63, e00543-e619. https://doi.org/10.1128/AAC.00543-19 (2019). Article PubMed PubMed Central Google Scholar * Loiko, V. & Wagener, J. The paradoxical effect of
echinocandins in _Aspergillus fumigatus_ relies on recovery of the β-1,3-glucan synthase Fks1. _Antimicrob. Agents Chemother._ 61, e01690-e1716. https://doi.org/10.1128/AAC.01690-16 (2017).
Article CAS PubMed PubMed Central Google Scholar * Tobin, M. B., Peery, R. B. & Skatrud, P. L. Genes encoding multiple drug resistance-like proteins in _Aspergillus fumigatus_ and
_Aspergillus flavus_. _Gene_ 200, 11–23. https://doi.org/10.1016/s0378-1119(97)00281-3 (1997). Article CAS PubMed Google Scholar * Slaven, J. W. _et al._ Increased expression of a novel
_Aspergillus fumigatus_ ABC transporter gene, atrF, in the presence of itraconazole in an itraconazole resistant clinical isolate. _Fungal Genet. Biol._ 36, 199–206.
https://doi.org/10.1016/s1087-1845(02)00016-6 (2002). Article CAS PubMed Google Scholar * Nascimento, A. M. _et al._ Multiple resistance mechanisms among _Aspergillus fumigatus_ mutants
with high-level resistance to itraconazole. _Antimicrob. Agents Chemother._ 47, 1719–1726. https://doi.org/10.1128/AAC.47.5.1719-1726.2003 (2003). Article CAS PubMed PubMed Central
Google Scholar * Poulsen, J. S., Madsen, A. M., White, J. K. & Nielsen, J. L. Physiological responses of _Aspergillus_ _niger_ challenged with itraconazole. _Antimicrob. Agents
Chemother._ 18, e02549-e2620. https://doi.org/10.1128/AAC.02549-20.PMID:33820768;PMCID:PMC8316071 (2021). Article Google Scholar * Gautam, P. _et al._ Proteomic and transcriptomic analysis
of _Aspergillus fumigatus_ on exposure to amphotericin B. _Antimicrob. Agents Chemother._ 52(12), 4220–4227. https://doi.org/10.1128/aac.01431-07 (2008). Article CAS PubMed PubMed
Central Google Scholar * da Silva Ferreira, M. E. _et al._ Transcriptome analysis of _Aspergillus fumigatus_ exposed to voriconazole. _Curr. Genet._ 50, 32–44.
https://doi.org/10.1007/s00294-006-0073-2 (2006). Article CAS PubMed Google Scholar * Fraczek, M. G. _et al._ The cdr1B efflux transporter is associated with non-cyp51a-mediated
itraconazole resistance in _Aspergillus fumigatus_. _J. Antimicrob. Chemother._ 68, 1486–1496. https://doi.org/10.1093/jac/dkt075 (2013). Article CAS PubMed Google Scholar * Balajee, S.
A. _et al._ Molecular identification of _Aspergillus_ species collected for the transplant-associated infection surveillance network. _J. Clin. Microbiol._ 47, 3138–3141.
https://doi.org/10.1128/JCM.01070-09 (2009). Article CAS PubMed PubMed Central Google Scholar * Frías-De-León, M. G. _et al._ Identification of _Aspergillus tubingensis_ in a primary
skin infection. _J. Mycol. Med._ 28, 274–278. https://doi.org/10.1016/j.mycmed.2018.02.013 (2018). Article PubMed Google Scholar * Asano, M., Kano, R., Makimura, K., Hasegawa, A. &
Kamata, H. Molecular typing and in-vitro activity of azoles against clinical isolates of _Aspergillus fumigatus_ and _A. niger_ in Japan. _J. Infect. Chemother._ 17, 483–6.
https://doi.org/10.1007/s10156-010-0202-1 (2011). Article PubMed Google Scholar * Hoda, S. _et al._ Inhibition of _Aspergillus_ _fumigatus_ bioflm and cytotoxicity study of natural
compound Cis9-hexadecenal. _J. Pure Appl. Microbiol._ 13, 1207–1216. https://doi.org/10.22207/JPAM.13.2.61 (2019). Article Google Scholar * Chowdhary, A., Sharma, C., Kathuria, S., Hagen,
F. & Meis, J. F. Azole-resistant _Aspergillus fumigatus_ with the environmental TR46/Y121F/T289A mutation in India. _J. Antimicrob. Chemother._ 69(2), 555–557.
https://doi.org/10.1093/jac/dkt397 (2014). Article CAS PubMed Google Scholar * Chowdhary, A. _et al._ Clonal expansion and emergence of environmental multiple-triazole-resistant
_Aspergillus fumigatus_ strains carrying the TR34/L98H mutations in the cyp 51A gene in India. _PloS One_ 7(12), e52871. https://doi.org/10.1371/journal.pone.0052871 (2012). Article ADS
CAS PubMed PubMed Central Google Scholar * Badali, H. _et al._ _In vitro_ activities of five antifungal drugs against opportunistic agents of _Aspergillus Nigri_ complex.
_Mycopathologia_ 181, 235–240. https://doi.org/10.1007/s11046-015-9968-0 (2016). Article CAS PubMed Google Scholar * Hashimoto, A. _et al._ Drug sensitivity and resistance mechanism in
_Aspergillus_ section _Nigri_ strains from Japan. _Antimicrob. Agents Chemother._ 61, e02583-e2616. https://doi.org/10.1128/AAC.02583-16 (2017). Article CAS PubMed PubMed Central Google
Scholar * Al Abdallah, Q., Ge, W. & Fortwendel, J. R. A simple and universal system for gene manipulation in _Aspergillus fumigatus_: In vitro-assembled Cas9-guide RNA
ribonucleoproteins coupled with microhomology repair templates. _Msphere_ 2(6), 10–1128. https://doi.org/10.1128/msphere.00446-17 (2017). Article CAS Google Scholar * Umeyama, T. _et al._
CRISPR/Cas9 genome editing to demonstrate the contribution of Cyp51A Gly138Ser to azole Resistance in _Aspergillus fumigatus_. _Antimicrob. Agents Chemother._ 62(9), e00894-e918.
https://doi.org/10.1128/AAC.00894-18 (2018). Article CAS PubMed PubMed Central Google Scholar * Eddouzi, J. _et al._ Molecular mechanisms of drug resistance in clinical _Candida_
species isolated from Tunisian hospitals. _Antimicrob. Agents Chemother._ 57, 3182–3193. https://doi.org/10.1128/AAC.00555-13 (2013). Article CAS PubMed PubMed Central Google Scholar *
Pérez-Cantero, A., Martin-Vicente, A., Guarro, J., Fortwendel, J. R. & Capilla, J. Analysis of the cyp51 genes contribution to azole resistance in Aspergillus section Nigri with the
CRISPR-Cas9 technique. _Antimicrob. Agents Chemother._ 1, e01996-e2020. https://doi.org/10.1128/AAC.01996-20 (2023). Article Google Scholar * Cannon, R. D. _et al._ Efflux-mediated
antifungal drug resistance. _Clin. Microbiol. Rev._ 22, 291–321. https://doi.org/10.1128/CMR.00051-08 (2009). Article CAS PubMed PubMed Central Google Scholar * Paul, R. A. _et al._
Magnitude of voriconazole resistance in clinical and environmental isolates of _Aspergillus flavus_ and investigation into the role of multidrug efflux pumps. _Antimicrob. Agents Chemother._
62, e01022-e1118. https://doi.org/10.1128/AAC.01022-18 (2018). Article PubMed PubMed Central Google Scholar * Sharma, C., Nelson-Sathi, S., Singh, A., Radhakrishna Pillai, M. &
Chowdhary, A. Genomic perspective of triazole resistance in clinical and environmental Aspergillus fumigatus isolates without cyp51A mutations. _Fungal. Genet Biol._ 26, 103265.
https://doi.org/10.1016/j.fgb.2019.103265 (2019). Article CAS Google Scholar * Sharma, C. _et al._ Investigation of multiple resistance mechanisms in voriconazole-resistant _Aspergillus
flavus_ clinical isolates from a chest hospital surveillance in Delhi, India. _Antimicrob. Agents Chemother._ 62, e01928-e2017. https://doi.org/10.1128/AAC.01928-17 (2018). Article PubMed
PubMed Central Google Scholar * Choi, M. J. _et al._ Microsatellite typing and resistance mechanism analysis of voriconazole-resistant _Aspergillus flavus_ Isolates in South Korean
hospitals. _Antimicrob. Agents Chemother._ 63, e01610-e1618. https://doi.org/10.1128/AAC.01610-18 (2019). Article CAS PubMed PubMed Central Google Scholar * Sen, P. _et al._
4-Allyl-2-methoxyphenol modulates the expression of genes involved in efflux pump, biofilm formation and sterol biosynthesis in azole resistant _Aspergillus fumigatus_. _Front. Cell. Infect.
Microbiol._ 13, 20. https://doi.org/10.3389/fcimb.2023.1103957 (2023). Article CAS Google Scholar * de Hoog, G., Guarr, J., Tran, C. S., Wintermans, R. G. F. & Gene, J. _Hyphomycetes
Atlas of Clinical Fungi_ (Wiley, 1995). Google Scholar * Alexander, B. D. Clinical and Laboratory Standards Institute. Reference method for broth dilution antifungal susceptibility testing
of filamentous fungi. 3rd ed. CLSI standard M38 (ISBN 1-56238-830-4) Pennsylvania USA, (2017). * Verweij, P. E., Howard, S. J., Melchers, W. J. & Denning, D. W. Azole resistance in
_Aspergillus_: Proposed nomenclature and breakpoints. _Drug Resist. Updat._ 12, 141–147. https://doi.org/10.1016/j.drup.2009.09.002 (2009). Article CAS PubMed Google Scholar *
Espinel-Ingroff, A. _et al._ Wild-type MIC distributions and epidemiological cutoff values for the triazoles and six Aspergillus spp. for the CLSI broth microdilution method (M38–A2
document). _J. Clin. Microbiol._ 48, 3251–7. https://doi.org/10.1128/JCM.00536-10 (2010). Article CAS PubMed PubMed Central Google Scholar * Lee, S. B., Milgroom, M. G. & Taylor, J.
W. A rapid, high yield mini-prep method for isolation of total genomic DNA from fungi. _Fungal Genet._ 35, 23–24. https://doi.org/10.4148/1941-4765.1531 (1988). Article Google Scholar *
Wu, Z. H., Wang, T. H., Huang, W. & Qu, Y. B. A simplified method for chromosome DNA preparation from filamentous Fungi. _Mycosystema_ 20, 575–577 (2001). CAS Google Scholar * White,
T. J., Bruns, T., Lee, S. & Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In _PCR Protocols a Guide to Methods and Applications_ (eds
Innis, M. A. _et al._) (Academic Press, 1990). Google Scholar * Mellado, E. _et al._ A new _Aspergillus fumigatus_ resistance mechanism conferring in vitro cross-resistance to azole
antifungals involves a combination of cyp51A alterations. _Antimicrob. Agents Chemother._ 51, 1897–1904. https://doi.org/10.1128/AAC.01092-06 (2007). Article CAS PubMed PubMed Central
Google Scholar * Freyd, T. _et al._ Ligand-guided homology modelling of the GABAB2 subunit of the GABAB receptor. _PLoS One_ 12, e0173889. https://doi.org/10.1371/journal.pone.0173889
(2017). Article CAS PubMed PubMed Central Google Scholar * Halgren, T. A. _et al._ Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database
screening. _J. Med. Chem._ 47, 1750–1759. https://doi.org/10.1021/jm030644s (2004). Article CAS PubMed Google Scholar * Harder, E. _et al._ OPLS3: A force field providing broad coverage
of drug-like small molecules and proteins. _J. Chem. Theory Comput._ 12, 281–296. https://doi.org/10.1021/acs.jctc.5b00864 (2016). Article CAS PubMed Google Scholar * Kamboj, H. _et al._
Gene expression, molecular docking, and molecular dynamics studies to identify potential antifungal compounds targeting virulence proteins/genes VelB and THR as possible drug targets
against _Curvularia lunata_. _Front. Mol. Biosci._ 9, 1055945. https://doi.org/10.3389/fmolb.2022.1055945 (2022). Article CAS PubMed PubMed Central Google Scholar * Gupta, L., Sen, P.,
Bhattacharya, A. K. & Vijayaraghavan, P. Isoeugenol affects expression pattern of conidial hydrophobin gene RodA and transcriptional regulators MedA and SomA responsible for adherence
and biofilm formation in Aspergillus fumigatus. _Arch. Microbiol._ 204, 214. https://doi.org/10.1007/s00203-022-02817-w (2022). Article CAS PubMed PubMed Central Google Scholar Download
references AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Amity Institute of Biotechnology, Amity University Uttar Pradesh, Sector-125, Noida, Uttar Pradesh, India Pooja Sen, Mukund Vijay,
Himanshu Kamboj, Lovely Gupta & Pooja Vijayaraghavan * Department of Biotechnology and Bioinformatics, Jaypee University of Information Technology, Solan, India Jata Shankar Authors *
Pooja Sen View author publications You can also search for this author inPubMed Google Scholar * Mukund Vijay View author publications You can also search for this author inPubMed Google
Scholar * Himanshu Kamboj View author publications You can also search for this author inPubMed Google Scholar * Lovely Gupta View author publications You can also search for this author
inPubMed Google Scholar * Jata Shankar View author publications You can also search for this author inPubMed Google Scholar * Pooja Vijayaraghavan View author publications You can also
search for this author inPubMed Google Scholar CONTRIBUTIONS P.S. performed literature search, conducted all the experiments, and drafted the manuscript; M.V. performed experiments and
assisted in manuscript writing; H.K. and L.G. performed the homology modeling and assisted in manuscript writing; J.S. critically reviewed and revised the manuscript; and P.V. conceptualized
the idea and critically analyzed the manuscript. CORRESPONDING AUTHOR Correspondence to Pooja Vijayaraghavan. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing
interests. ADDITIONAL INFORMATION PUBLISHER'S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY
INFORMATION SUPPLEMENTARY INFORMATION. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing,
adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons
licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise
in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the
permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Sen, P., Vijay, M., Kamboj, H. _et al._ _cyp51A_ mutations, protein modeling, and efflux pump gene expression reveals multifactorial
complexity towards understanding _Aspergillus_ section _Nigri_ azole resistance mechanism. _Sci Rep_ 14, 6156 (2024). https://doi.org/10.1038/s41598-024-55237-9 Download citation * Received:
18 October 2023 * Accepted: 21 February 2024 * Published: 14 March 2024 * DOI: https://doi.org/10.1038/s41598-024-55237-9 SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative