Play all audios:
ABSTRACT _Helicobacter pylori_ infection is a major risk factor for gastric adenocarcinomas. In the case of the intestinal subtype, chronic gastritis and intestinal metaplasia are well-known
sequential steps in carcinogenesis. _H. pylori_ has high genetic diversity that can modulate virulence and pathogenicity in the human host as a cag Pathogenicity Island (cagPAI). However,
bacterial gene combinations do not always explain the clinical presentation of the disease, indicating that other factors associated with _H. pylori_ may play a role in the development of
gastric disease. In this context, we characterized the microbial composition of patients with chronic gastritis (inactive and active), intestinal metaplasia, and gastric cancer as well as
their potential association with _H. pylori_. To this end, _16 S_ rRNA metagenomic analysis was performed on gastric mucosa samples from patients with different types of lesions and normal
gastric tissues. Our main finding was that _H. pylori_ virulence status can contribute to significant differences in the constitution of the gastric microbiota between the sequential steps
of the carcinogenesis cascade. Differential microbiota was observed in inactive and active gastritis dependent of the _H. pylori_ presence and status (_p_ = 0.000575). _Pseudomonades_, the
most abundant order in the gastritis, was associated the presence of non-virulent _H. pylori_ in the active gastritis. Notably, there are indicator genera according to _H. pylori_ status
that are poorly associated with diseases and provide additional evidence that the microbiota, in addition to _H. pylori_, is relevant to gastric carcinogenesis. SIMILAR CONTENT BEING VIEWED
BY OTHERS META-ANALYSIS OF MUCOSAL MICROBIOTA REVEALS UNIVERSAL MICROBIAL SIGNATURES AND DYSBIOSIS IN GASTRIC CARCINOGENESIS Article Open access 09 June 2022 MICROBIAL COMPOSITION OF
TUMOROUS AND ADJACENT GASTRIC TISSUE IS ASSOCIATED WITH PROGNOSIS OF GASTRIC CANCER Article Open access 21 March 2023 HUMAN GASTRIC MICROBIOTA ANALYSIS OF REFRACTORY _H. PYLORI_ INFECTION
Article Open access 07 July 2024 INTRODUCTION Gastric cancer (GC) is a global public health problem owing to its high incidence and mortality rates. It is the fifth most frequently diagnosed
cancer and the fourth leading cause of cancer-related death according to the Globocan estimative1. Approximately 95% of gastric tumors are adenocarcinomas that can histologically be
classified as diffuse or intestinal type2. _Helicobacter pylori_ infection is the major risk factor for gastric adenocarcinomas in which, chronic active gastritis and intestinal metaplasia
are part of bacterium-related intestinal subtype3. Notably, _H. pylori_ has co-evolved with humans up to approximately l00 years ago4,5, indicating a bacterium-host equilibrium, and this
symbiotic relationship must be altered for the GC development. _H. pylori_ has high genetic diversity due to rearrangements and horizontal gene transfer, which reflect historical human
demographic events. Among the horizontally acquired gene regions, cag pathogenicity island (cagPAI), a cluster of genes encoding the Type IV secretion system, contains a virulent factor that
mediates colonization and pathogenicity in the human host6,7. However, the island is not a uniformly conserved entity, and the correlation between different cagPAI rearrangements and
clinical presentation is not always found8,9, indicating that other factors, in association with _H. pylori_, play a role in the development of gastric disease. Currently, research using _16
S_ rRNA sequence analysis, and more recently, high-throughput sequencing technology, has revealed that the stomach harbors a distinct and complex ecosystem in addition to _H. pylori._ Under
physiological conditions, the microbiota plays an essential role in human health, and alterations in the microbiome have been hypothesized to be linked to gastric disease development, as
they can stimulate the inflammatory process10,11. Previous studies have investigated the characteristics of gastric microbiota in different gastric diseases; however, few studies have
addressed bacterial genotypes, which are relevant factors for GC development. In addition, considering the multifactorial nature of gastric lesions, diverse populations with different habits
should be studied to clarify this association. To address this issue, we characterized the microbial composition of patients with chronic gastritis, intestinal metaplasia, and GC. Given the
role of reactive oxygen species derived from neutrophils, which are characteristic of ACG and linked to DNA damage, the gastritis was categorized as inactive (ICG) or active (ACG), In
addition, we explored potential associations with _H. pylori_ strains, considering the cagPAI genes, _cagA_ and _cagE_, which have not yet been considered in gastric microbiota studies.
PATIENTS AND METHODS Sixty-six patients were included in this study after selection based on histological classification, gastric region, and _H. pylori_ genotype. Fifteen intestinal GC
samples were selected from 172 patients who underwent gastrectomy after GC diagnosis at the Hospital Universitario Walter Cantídeo (HUWC) and Santa Casa de Misericordia Hospital in
Fortaleza, Ceará, Brazil. Forty-eight non-cancerous gastric lesions (NGL), including 20 intestinal metaplasia, 12 active chronic gastritis, 16 inactive chronic gastritis, and 3 normal
gastric tissues, were selected from 202 patients with dyspeptic symptoms who underwent endoscopy at the HUWC and Hospital Geral de Fortaleza, Ceará, Brazil. All patients were from a
non-cardiac region. Exclusion criteria for NGL were the presence of duodenal gastric or esophageal masses, Barrett’s esophagus lesions, previous gastric or duodenal surgery, treatment with
any anti-inflammatory, proton-pump inhibitor drugs, or antibiotics during the last three months and previous therapy for _H. pylori._ The histopathological diagnosis of GC was made using
Lauren12 and NGL according to the update Sydney system13. ICG was defined as diffuse lymphocytic inflammatory infiltration or organization in the follicular/nodular structures. ACG was
defined by the presence of neutrophilic infiltrates permeating the glands and/or lamina propria. Normal cases included patients who underwent endoscopy with negative results and were _H.
pylori_ negative. Table 1 presents the demographic data of the study participants grouped according to the type of gastric lesion. DNA EXTRACTION, _H. PYLORI_DETECTION AND GENOTYPING
Metagenomic DNA was extracted from frozen tumor tissue samples consisting mainly of tumor cells (> 80%) and single frozen stomach fragments in which the NGL was representative, using the
cetyltrimethylammonium bromide method adapted from Foster and Twell14. Concentration and quality of metagenomic DNA were determinate with NanoDrop ND-1000®. _H. pylori_ infection was
detected using amplification of the _ureC_ gene as described by Lage et al.15. cagPAI genes (_cagA_ and _cagE_) were identified using specific primers and conditions described by Lima et
al.16. Negative (water) and positive controls were assayed in each run. The PCR products were visualized using 1% agarose gel electrophoresis with ethidium bromide. 16SRRNA AMPLICON FOR
LIBRARY PREPARATION For 16 S rRNA gene amplification, DNA from all samples selected for this study was purified using DNA clean-up kit (Qiagen, Germany). The amplicon library of the V4
variable region of the 16 S rRNA gene was prepared according to the protocol suggested by Illumina (San Diego, CA, USA), using primers 515 F/806R.17 PCR mixtures (25 µL) were composed of Taq
HotStart HiFi buffer plus, 1U of Taq HotStar High Fidelity (Kapa Biosystems, Boston, MA, EUA), 0.75mM dNTPs, and 0.75mM of each primer and 30 ng of DNA template of each sample. PCR
amplification was carried out under the following conditions: 94°C for 4 min of initial heating followed 25 cycles of 94°C for 45 s, 60°C for 60 s and 72°C for 60 s, after which a final
elongation step at 72°C for 10 min. Each sample was put in triplet with one negative control. The success of the amplification, with expected bands around 250 bp, was assessed on a 1%
agarose gel. Library preparation was done using Illumina Nextera XT (Illumina, San Diego, CA, USA), according to the manufacturer’s instructions. Briefly, 5 µL of the each confirmed
amplified sample was submitted to a second reaction in a 50 µL PCR mixture using 1x Taq HotStart HiFi buffer, 1U of Taq DNA polimerase HotStart High fidelity, (Kapa Biosystems, Boston, MA,
EUA), 0.75mM dNTPs and 5 µL of each Illumina Nextera XT index. PCR conditions consisted of initial heating of 94°C for 4 min followed 8 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 30
s and final elongation steps at 72°C for 5 min. The PCR products were purified using Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, CA, USA) according to the manufacturer’s
instructions. Amplicons from each sample were pooled and quantified by spectrophotometry using a dsDNA BR Assay Kit (Invitrogen). DNA libraries were multiplexed using equal amount of the
pooled amplicon, diluted to a final concentration of 8.0 pM and it was spiked with the PhiX sequencing positive control (Illumina), in a final ratio of 80:20. Sequencing was performed on an
Illumina MiSeq platform (Illumina, San Diego, CA, USA) in a 2 × 300 paired-end configuration. SEQUENCING DATA PROCESSING After sequencing, Illumina adapter sequences were trimmed from
already-demultiplexed raw fastq files using Cutadapt v1.818 in paired-end mode, and the reads quality was assessed using FastQC v.0.11.819 and vsearch v2.10.420. Subsequent analyses were
performed in the R v3.5.3 environment21, following the DADA2 v1.11.1 package22 pipeline for obtaining a table of non-chimeric amplicon sequence variants (ASVs) free of low-quality and
non-prokaryotic sequences (sequences differing by as little as one nucleotide)23. Taxonomy assignment and removal of non-prokaryotic sequences was performed against the SILVA reference
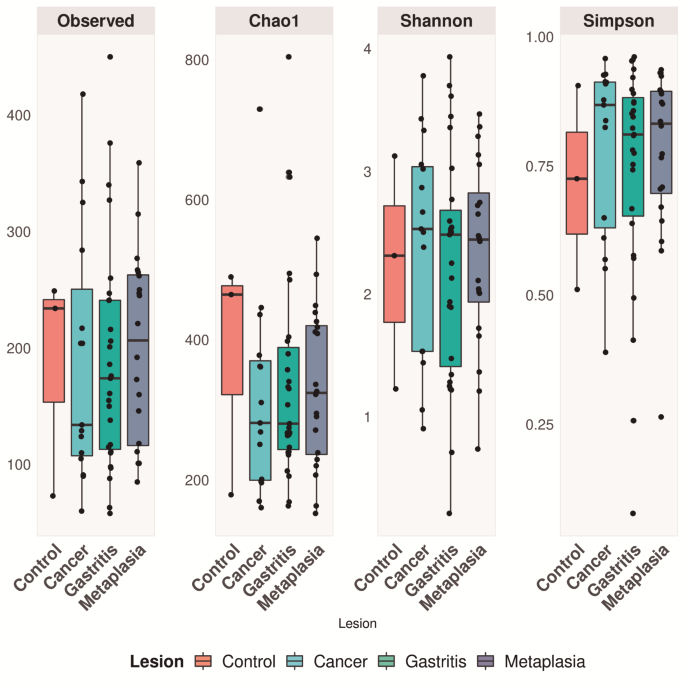
database (release 132)24. DATA ANALYSES Observed, Chao1, Shannon and Simpson were calculated for alpha diversity evaluation. Alpha diversity metrics summarize the structure of an ecological
community with respect to its richness (number of taxonomic groups) and evenness (distribution of abundances of the groups). Shapiro-Wilk test was used to assess whether the data set was
normally distributed. As Shannon diversity was parametric, one-way analysis of variance and Tukey’s honest significant difference (HSD) post-hoc tests were used for multiple comparisons of
means at a 95% confidence interval. For Chao1 and observed ASVs, we used the Kruskal–Wallis non-parametric test25. To analyze beta diversity, microbial communities were compared using
distance-based comparisons and a comparison of composition profiles related to each of the studied lesions. For the distance-based approach, the relative abundances of ASVs were square-root
transformed, and the Bray-Curtis similarity coefficient was calculated for each sample pair. The similarity matrix was visualized using nonmetric multidimensional scaling. Indicator analysis
was performed using the package _indicspecies_26 to identify the main genera associated with each type of lesion, clinical status, and virulence. The relationship between clinical variables
and the bacterial community was evaluated through canonical correspondence analysis (CCA), using the Vegan package26 in the R environment21. The ‘decorana’ function was used to choose the
best ordering model, which indicated the use of CCA. Preliminary CCA was performed using all available variables (age, lesion type, cagA, cagE, virb11, and other genotypic markers). The
variables that demonstrated significant influence (p < 0.05) were selected using the ‘ordistep’ function and confirmed using the ‘anova.cca’ function for the final CCA. The inflation
factor of the variation (function ‘vif.cca’) showed that the retained explanatory variables did not present collinearity among themselves (FIV < 10). In addition, the samples were
classified into 11 groups, considering gastric lesions and _H. pylori_ genes cagA/cagE, as described in Table 2. RESULTS SEQUENCING AND COVERAGE DATA Metagenomic data collected from 66
samples were grouped according to histopathological classification, _H. pylori_, and virulence status. From these, 1,957,697 high-quality reads, with a minimum of 125 sequences (CG150
sample) and a maximum of 72,056 sequences (LG76 sample), were obtained. Three samples had fewer than 10,000 sequences (CG150, LG171, and TG10) and were excluded from subsequent analyses. The
Good’s coverage estimator was > 97% for the overall sequence set. ALPHA DIVERSITY Alpha diversity was evaluated using Chao1-richness, Simpson-diversity index, and Shannon diversity index
(combining richness and diversity). The Shapiro-Wilk test showed that all diversity metrics for this dataset were non-parametric, except for the Shannon test. ANOVA followed by Tukey’s test
for Shannon and Kruskal–Wallis were applied to the other estimators, but no statistically significant difference was observed between them for the alpha diversity metric (Fig. 1).
Nevertheless, it was possible to observe from Fig. 2, a microbial signature composition in each lesion group. GLOBAL TAXONOMIC COMPOSITION PHYLUM Operational taxonomic units (OTUs) with >
5% abundance were affiliated with 10 bacterial phyla. The phylum composition varied among the different gastric lesions (_p_ = 0.0292, two-way ANOVA and Tukey’s HSD test) (Fig. 2). A
statistically significant difference in microbial composition was observed in each group according to the inflammation status (_p_ = 1.26e-11), _H. pylori_ presence (_p_ = 0.000133), and
virulence (_p_ = 0.044126). In the metaplasia group, the microbial composition differed from that of the control (_p_ = 6.73e-09) and cancer groups (_p_ = 0.049779) (Fig. 3). The abundance
of the phylum _Firmicutes_ in the control cluster decreased among the gastric lesions; conversely, _Actinobacteria_ were present in all gastric lesions but not in normal tissue. Notably, in
addition to _Firmicutes_ being the most abundant phylum in the control group, this was also the case for inactive gastritis without _H. pylori_ and non-virulent _H. pylori_. (_p_ = 2.11
e−12). The _Proteobacteria_ phylum was the most frequently detected group in cancer, metaplasia, and active gastritis. Notably, we observed a negative association between the presence of
_Firmicute_s and _Helicobacter_ spp. (_p_ = 1.45e-07) in this group (Fig. 3), with _Helicobacter_ spp. being statistically more abundant in the metaplasia samples than in the other lesions
(Fig. 2). No significant differences in _H. pylori_ virulence were observed in the cancer group (Fig. 3). ORDER At the order level, the abundance significantly varied among lesions (_p_ =
3.3e-09). The most frequent order in the control group was _Bacillales_, while that in gastritis was _Pseudomonadales_ and _Campylobacterales_ and, in metaplasia was _Campylobacterales_
(Fig. 4). Between gastritis, the _H. pylori_ presence and status significantly changed the microbiota composition (_p_ = 0.000575), with _Pseudomonadales_ being more abundant in active
gastritis carrying non-virulent _H. pylori_ strains (Fig. 5). In metaplasia, the microbial composition differed significantly from that of the control (_p_ = 2.15e-08) and between the
presence and absence of _H. pylori_ (6.392 3.4e-16). In addition, a negative association was observed between _Bacillales_ and _Campylobacterales_ (Fig. 6). Considering the cancer group,
_Sphingomonadales_ was more abundant than that in the control group (_p_ = 0.00391) (Fig. 4). GENUS A diverse community of 270 genera was detected in this study. Among these, 79, 205, 176,
and 177 genera were in controls, gastritis, metaplasia, and cancer patients, respectively. Overall, _Helicobacter_ was the most common genus, followed by _Pseudomonas_ and _Bacillus_.
Analyses at the genus level (abundance > 20%) are presented in Figs. 7, 8, 9 and 10. A significant difference in microbiome abundance was observed in the general comparison between
lesions (_p_ = 2e-16), Fig. 7). Gastritis status (active and non-active) still showed differences in microbiome composition, but there was a notable positive association between the presence
of _Pseudomonas_ and _Helicobacter_, especially in active gastritis (Fig. 8). A higher frequency of _Bacillus_ was observed in the control and inactive gastritis groups (Fig. 8). Similarly,
among the metaplasia clusters, the differential microbiomes were influenced by _H. pylori_ infection (_p_ = 2.67e-09; Fig. 9). The microbial composition differed between the control and
cancer groups (_p_ = 0.0045), with _Sphingomonas_ being more frequent in the cancer group. Although there was no significant difference in virulence, Fig. 10 shows a greater abundance of
this genus in the non-virulent _H. pylori_ cluster. Finally, Fig. 11 summarizes the main genera involved in each lesion group classified into families. INDICATOR GENUS Indicator analysis was
performed to identify the main genera associated with lesion type, clinical status, and virulence. This analysis revealed that 10 of 270 genera were responsible for the statistically
significant differences. Most indicator genera were part of a rare biosphere, with abundances below 0.01%. Four of them (_Microvirga_,_ Massilia_,_ Mesorhizobium_, and _Aquincola_) were
associated with the control group, _Salinicola_ was associated with active gastritis, _Lactobacillus_ was associated with the cancer group, _Raoultella_ and _Leuconostoc_ were associated
with cancer non-virulent _H. pylori_ and _Sarcina_ and _Centipeda_ were associated with cancer virulent HP (Table 3; Figs. 11 and 12). DISCUSSION In this study, we investigated the gastric
microbiota by considering sequential histological events in intestinal GC development. Gastritis was grouped according to neutrophilic infiltration as ACG or ICG, reflecting differences in
host response to infections. In addition, _H. pylori_ was categorized as virulent mainly by the presence of the _cagA_ or _cagE_ genes. This aspect differentiates the present study from the
gastric metagenomics currently published, in which the association with _H. pylori_ genotype, when performed, was only with the presence of cagA. Consistent with previous studies, the phylum
composition differed among the gastric lesion’s studies27,28,29,30; however, Dicksved31 and Wang et al.32 did not find any differences in the diversity indices of bacterial phyla.
Nonetheless, both studies had a small number of samples; the study by Dicksved included 10 patients with GC and 5 controls with dyspepsia, while in Wang’s study, only 12 out of 315 patients
with gastritis and GC were not analyzed using _16 S_ rRNA NGS. _Firmicutes_ seem to be associated with more benign lesions since this phylum was statistically more abundant in the control
group and with inactive gastritis in the absence or presence of less virulent _H. pylori_. In a study by Gantuya et al.33 with gastritis patients from Mongolia, an area with high GC
incidence and mortality, Firmicutes was the most frequent phylum in the control group and was negative _for H. pylori._ Thus, a negative association between the presence of _Firmicutes_ and
_Helicobacter spp._ was observed. Consistent with this result, Maldonado-Contreras et al.34 in a study aiming to characterize the structure of the human gastric bacterial community in
relation to _H. pylori_ status, observed a decreased abundance of _Firmicutes_, _Actinobacteria_ and _Bacteroidetes_ in _H. pylori_ positive cases. In contrast to _Firmicutes_, the
_Actinobacteria_ phylum was found only in the lesions (not in the control samples); however, there were no differences among the groups with respect to _H. pylori_ status. The
_Proteobacteria_ phylum, which includes _Helicobacter_, was the most frequent group associated with cancer, metaplasia, and active gastritis. At order level, _Bacillales_ was the
_Firmicutes_ phylum in the control group. _Bacillales_ play an active role in the production of bioproducts, such as organic acids, chemicals, surfactants, enzymes, and insecticides35. Also,
_Bacillales_ are found in milk and cheese36, and have been used in the food industry35. _Staphylococci_, _a_ representative of _Bacillales_, are abundant in the human skin microbiome, with
_Staphylococcus epidermidis_ being the most frequently isolated species37,38. This explains the predominance of the order in the control group. _Protobacteria_ were represented by
_Pseudomonadales_, the most abundant order in gastritis; however, their abundance was associated with the presence of non-virulent _H. pylori_ in active gastritis. The association
_Proteobacteria_, including _Pseudomonadales_, with the presence of inflammation has been observed in pediatric Crohn disease patients39 which may explain gastric activity in the presence of
less virulent _H. pylori_. In contrast, _Campylobacterales_, the order in which _H. pylori_ belongs, was the most abundant order in metaplasia, which is in accordance with the Correia
cascade for intestinal cancer progression, associated with _H. pylori_ as the main etiologic factor. A diverse community of 270 genera was detected in this study. Among these, 79, 205, 176
and 177 genera were in controls, gastritis, metaplasia and cancer patients, respectively. Overall, _Helicobacter_ was the most common genus, followed by _Pseudomonas_ and _Bacillus._ In this
study, ten genera were highlighted because they were related to the groups of lesions. Most of these indicator genera were part of the rare microbiota with abundances below 0.01%.
_Lactobacillus_, which was associated with cancer samples is in accordance with other studies40,41,42,43. The higher relative _Lactobacillus_ abundance in cancer could be a consequence of
its overgrowth as a result of the metabolic changes that occur during the carcinogenesis process, in which the pH increases due to the gradual reduction in gastric secretion. This gastric
acid reduction can lead to colonization of the stomach by species from ingestion, migration from the oropharyngeal cavity or by enterogastric reflux44,45,46. In this scenario,
_Lactobacillus_, being lactic acid bacteria, could tolerate this new acid level and proliferate, increasing their relative abundance47,48. In addition, the metabolism of oral _Lactobacillus_
generates volatile sulphur compounds, short-chain fatty acids (SCFAs), reactive oxygen species, hydrogen peroxide and lactic acid, all of which have already been implicated in chronic
inflammation, genomic instability and carcinogenesis49. Although species of this genus are widely used as probiotic microorganisms, it is important to highlight that biological behaviors
substantially vary between different species. Therefore, studies evaluating species that are effectively associated with GC are important for developing a therapeutic approach. In this
series of patients, it was also possible to identify the indicator genera according to _H. pylori_ status. An interesting association was found between _Leuconostoc_ and _Raoultella_ with
cancer-associated non-virulent _H. pylori. Leuconostoc spp_ are used in the food industry for their fermentation properties and production and contribute to the flavor of fermented
products50. They are not part of the human microbiota51. However, studies have reported infection by this microorganism in the blood, parenteral nutrition catheters, cerebrospinal fluid, and
urine, especially in immunocompromised patients52,53,54,55. Recently, Nouri et al.56 compared the oral microbiota of healthy individuals to patients with several cancer types (oral,
gastric, head and neck, and pancreatic) and observed that a higher abundance of _Leuconostoc_ was associated with a higher risk of all cancers. In addition, the authors suggested that
fluctuations in certain genera of oral bacteria, such as _Streptococcus_,_ Haemophilus_,_ Neisseria_, and _Leuconostoc_, could induce the production of SCFAs, causing the production of
cytokines and inflammation, potentially leading to cancer. Reports of human infections by the genus _Raoultella_ have increased over the last decade56. _Raoultella spp_. are very similar to
_Klebsiella_, sharing ecological, biochemical, clinical, and microbiological characteristics and making differential diagnosis very difficult with traditional microbiological techniques57.
These bacteria are found in plant ecosystems, water and soil and can colonize humans and animals57. _Raoultella spp_. can cause a wide range of clinical syndromes, such as bacteremia,
pneumonia, and urinary and biliary tract infections, especially in immunocompetent patients58. Except for a case report of a patient with early GC and _Raoultella planticola_ bacteremia59,
our study is the first to report _Raoultella_ spp. as an indicator of GC. Further studies are needed to clarify its relationship with nonvirulent _H. pylori_ cancer samples. _Sarcina and
Centipeda_ were associated with cancer-virulent _H. pylori. Sarcina spp._ are present in soil and cereal grains60 and are often associated with delayed gastric emptying and movement of food
to the intestine in addition to dyspepsia, gastritis, ulcers, and gastric perforation61,62,63. Studies indicate _Sarcina ventriculi_ is a relevant species in gastric pathologies, as
extensively reviewed by Tartaglia et al.64. However, it should be noted that _Sarcina spp_. have also been detected in healthy individuals65. In our study, we observed an interesting
association of _Sarcina_ with the cancer-virulent _H. pylori_ group. The coexistence between _Sarcina_ and _H. pylori_ has already been reported by Sauter et al. in patients with
Gastritis/Duodenitis66. Only one study, from Ugarde et al. (2022), reported infection by _Sarcina ventriculi_ in the biopsy of two gastric tumors67. These data encouraged us to conduct more
in-depth analyses of this notable relationship between virulent _H. pylori_ and _Sarcina ventriculi_. Regarding _Centipeda_, the most relevant findings were its association with chronic
periodontitis68, dental caries69 and, as a driver for colorectal carcinoma70 and its prognostic value for patients with oral cancer71. Our study is the first to report this genus in gastric
tumor samples, and this relationship should be studied in greater depth. The genus _Salinicola_, classified in the _Halomonadaceae_ family, currently includes 12 gram-negative, aerobic or
facultative anaerobes, which are halophilic. It is widely distributed in saline aquatic and terrestrial habitats and is associated with halophyte plants and sea animals. _Salinicola_ species
or strains have never been isolated from unequivocally pathological material from humans, animals, or plants72. This species was identified in our study as an indicator of active gastritis
(_p_ = 0.001), therefore, studies that clarify this relationship will be relevant. Overall, this series of patients demonstrated that _H. pylori_ virulence status can contribute to
significant differences in the composition of the gastric microbiota between the sequential steps of the carcinogenesis cascade. The identification of indicative bacterial genera is
emphasized, which are still poorly associated with diseases, and provides additional evidence that the microbiota is relevant for gastric carcinogenesis in addition _H. pylori_. Further
studies with functional analysis are needed to clarify the clinical implications of these findings. DATA AVAILABILITY The datasets generated and/or analyzed during the current study are
available in the National Center for Biotechnology Information repository at [https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1145484] (accession number PRJNA1145484). Additional information is
available from the corresponding author upon request. REFERENCES * Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in
185 countries. _CA Cancer J Clin._ 71(3), 209–249. https://doi.org/10.3322/caac.21660 (2021). Article CAS PubMed Google Scholar * Oue, N., Sentani, K., Sakamoto, N., Uraoka, N. &
Yasui, W. Molecular carcinogenesis of gastric cancer: Lauren classification, mucin phenotype expression, and cancer stem cells. _Int J Clin Oncol._ 24(7), 771–778.
https://doi.org/10.1007/s10147-019-01443-9 (2019). Article CAS PubMed Google Scholar * Conteduca, V. et al. pylori infection and gastric cancer: state of the art (review). _Int J Oncol._
42(1), 5–18. https://doi.org/10.3892/ijo.2012.1701 (2013). Article CAS PubMed Google Scholar * Moodley, Y. et al. Age of the association between Helicobacter pylori and man. _PLoS
Pathog._ 8(5), e1002693. https://doi.org/10.1371/journal.ppat.1002693 (2012). Article CAS PubMed PubMed Central Google Scholar * Maixner, F. et al. The 5300-year-old Helicobacter pylori
genome of the Iceman. _Science._ 351(6269), 162–165. https://doi.org/10.1126/science.aad2545 (2016). Article ADS CAS PubMed PubMed Central Google Scholar * Olbermann, P. et al. A
global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. _PLoS Genet._ 6(8), e1001069. https://doi.org/10.1371/journal.pgen.1001069
(2010). Article CAS PubMed PubMed Central Google Scholar * Kennemann, L. et al. Helicobacter pylori genome evolution during human infection. _Proc Natl Acad Sci U S A._ 108(12),
5033–5038. https://doi.org/10.1073/pnas.1018444108 (2011). Article ADS PubMed PubMed Central Google Scholar * Nishiya, D. et al. Genes inside the cagPAI of Helicobacter pylori are not
associated with gastric cancer in Japan. _Hepatogastroenterology._ 51(57), 891–894 (2004) (PMID: 15143941). CAS PubMed Google Scholar * Matteo, M. J. et al. Helicobacter pylori cag
pathogenicity island genotype diversity within the gastric niche of a single host. _J Med Microbiol._ 56(Pt 5), 664–669. https://doi.org/10.1099/jmm.0.46885-0 (2007). Article CAS PubMed
Google Scholar * Panebianco, C., Potenza, A., Andriulli, A. & Pazienza, V. Exploring the microbiota to better understand gastrointestinal cancers physiology. _Clin Chem Lab Med._ 56(9),
1400–1412. https://doi.org/10.1515/cclm-2017-1163 (2018). Article CAS PubMed Google Scholar * Ortigão, R., Pimentel-Nunes, P., Dinis-Ribeiro, M. & Libânio, D. Gastrointestinal
microbiome - What we need to know in clinical practice. _GE Port J Gastroenterol._ 27(5), 336–351. https://doi.org/10.1159/000505036 (2020). Article PubMed PubMed Central Google Scholar
* Lauren, P. THE two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. _Acta Pathol Microbiol
Scand._ 64, 31–49 (1965). Article CAS PubMed Google Scholar * Dixon, M. F., Genta, R. M., Yardley, J. H. & Correa, P. Classification and grading of gastritis. The updated Sydney
System. International workshop on the histopathology of gastritis, Houston 1994. _Am J Surg Pathol._ 20(10), 1161–1181 (1996). Article CAS PubMed Google Scholar * Foster, G. D. &
Twell, D. _Plant Gene Isolation: Principles and Practice_ 426 (Wiley, 1996). Google Scholar * Lage, A. P. et al. Diagnosis of Helicobacter pylori infection by PCR: comparison with other
invasive techniques and detection of cagA gene in gastric biopsy specimens. _J Clin Microbiol._ 33(10), 2752–2756. https://doi.org/10.1128/jcm.33.10.2752-2756.1995 (1995). Article CAS
PubMed PubMed Central Google Scholar * Lima, V. P., Silva-Fernandes, I. J., Alves, M. K. & Rabenhorst, S. H. Prevalence of Helicobacter pylori genotypes (vacA, cagA, cagE and virB11)
in gastric cancer in Brazilian’s patients: An association with histopathological parameters. _Cancer Epidemiol._ 35(5), e32–e37. https://doi.org/10.1016/j.canep.2011.02.017 (2011). Article
CAS PubMed Google Scholar * Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. _Proc Natl Acad Sci USA_ 108(Suppl 1), 4516–4522.
https://doi.org/10.1073/pnas.1000080107 (2011). Article ADS PubMed Google Scholar * Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. _EMBnet. J._ 17,
10. https://doi.org/10.14806/ej.17.1.200 (2011). Article Google Scholar * Andrews, S. FastQC A quality control tool for high throughput sequence data. _Babraham Bioinformatics_.
http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2010). * Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: A versatile open source tool for metagenomics.
_PeerJ._ 4, e2584. https://doi.org/10.7717/peerj.2584.PMID:27781170;PMCID:PMC5075697 (2016). Article PubMed PubMed Central Google Scholar * R Core Team. _R: The R Project for Statistical
Computing_. (R core team, 2016). * Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. _Nat Methods._ 13(7), 581–3.
https://doi.org/10.1038/nmeth.3869 (2016). Article CAS PubMed PubMed Central Google Scholar * Yilmaz, P. et al. The SILVA and “All-species Living Tree Project (LTP)” taxonomic
frameworks. _Nucleic Acids Res._ 42, D643-8. https://doi.org/10.1093/nar/gkt1209 (2014). Article CAS PubMed Google Scholar * de Cáceres, M., Sol, D., Lapiedra, O. & Legendre, P. A
framework for estimating niche metrics using the resemblance between qualitative resources. _Oikos._ 120, 1341–1350. https://doi.org/10.1111/j.1600-0706.2011.19679.x (2011). Article ADS
Google Scholar * Tavares, T. C. L., Bezerra, W. M., Normando, L. R. O., Rosado, A. S. & Melo, V. M. M. Brazilian semi-arid mangroves-associated microbiome as pools of richness and
complexity in a changing world. _Front Microbiol._ 12, 715991. https://doi.org/10.3389/fmicb.2021.715991 (2021). Article PubMed PubMed Central Google Scholar * Oksanen, J. et al.
_Package “vegan” - Community Ecology Package_. 296 (2019). * Nardone, G. & Compare, D. The human gastric microbiota: Is it time to rethink the pathogenesis of stomach diseases. _United
Eur. Gastroenterol. J._ 3, 255–260. https://doi.org/10.1177/2050640614566846 (2015). Article CAS Google Scholar * Ferreira, R. M. et al. Gastric microbial community profiling reveals a
dysbiotic cancer-associated microbiota. _Gut._ 67, 226–236. https://doi.org/10.1136/gutjnl-2017-314205 (2017). Article CAS PubMed Google Scholar * Stewart, O. A., Wu, F. & Chen, Y.
The role of gastric microbiota in gastric cancer. _Gut Microbes._ 11, 1220–1230 (2020). Article CAS PubMed PubMed Central Google Scholar * Eun, C. S. et al. Differences in gastric
mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. _Helicobacter._ 19(6), 407–416.
https://doi.org/10.1111/hel.12145 (2014). Article CAS PubMed Google Scholar * Dicksved, J. et al. Characterization of the stomach microbiota in patients with gastric cancer and in
controls. _J Med Microbiol._ 58(4), 509–516. https://doi.org/10.1099/jmm.0.007302-0 (2009). Article CAS PubMed Google Scholar * Wang, L. et al. Bacterial overgrowth and diversification
of microbiota in gastric cancer. _Eur J Gastroenterol Hepatol._ 28(3), 261–266. https://doi.org/10.1097/MEG.0000000000000542 (2016). Article CAS PubMed PubMed Central Google Scholar *
Gantuya, B. et al. Gastric microbiota in helicobacter pylori-negative and -positive gastritis among high incidence of gastric cancer area. _Cancers (Basel)._ 11(4), 504.
https://doi.org/10.3390/cancers11040504 (2019). Article CAS PubMed PubMed Central Google Scholar * Maldonado-Contreras, A. et al. Structure of the human gastric bacterial community in
relation to Helicobacter pylori status. _ISME J._ 5(4), 574–579. https://doi.org/10.1038/ismej.2010.149 (2011). Article CAS PubMed Google Scholar * Harirchi, S. et al. Bacillales: From
taxonomy to biotechnological and industrial perspectives. _Microorganisms._ 10(12), 2355. https://doi.org/10.3390/microorganisms10122355 (2022). Article CAS PubMed PubMed Central Google
Scholar * Delbès, C., Ali-Mandjee, L. & Montel, M. C. Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analyses. _Appl
Environ Microbiol._ 73(6), 1882–1891. https://doi.org/10.1128/AEM.01716-06 (2007). Article ADS CAS PubMed PubMed Central Google Scholar * Coates, R., Moran, J. & Horsburgh, M. J.
Staphylococci: Colonizers and pathogens of human skin. _Future Microbiol._ 9(1), 75–91. https://doi.org/10.2217/fmb.13.145 (2014). Article CAS PubMed Google Scholar * Alonzo, F. Toward
uncovering the complexities of bacterial interspecies communication and competition on the skin. _mBio._ 13(4), e0132022. https://doi.org/10.1128/mbio.01320-22 (2022). Article PubMed
Google Scholar * Haberman, Y. et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. _J Clin Invest._ 124(8), 3617–3633.
https://doi.org/10.1172/JCI75436 (2014). Article CAS PubMed PubMed Central Google Scholar * Vinasco, K., Mitchell, H. M., Kaakoush, N. O. & Castaño-Rodríguez, N. Microbial
carcinogenesis: Lactic acid bacteria in gastric cancer. _Biochim Biophys Acta Rev Cancer._ 1872(2), 188309. https://doi.org/10.1016/j.bbcan.2019.07.004 (2019). Article CAS PubMed Google
Scholar * Li, Z. P. et al. Overgrowth of Lactobacillus in gastric cancer. _World J Gastrointest Oncol._ 13(9), 1099–1108. https://doi.org/10.4251/wjgo.v13.i9.1099 (2021). Article PubMed
PubMed Central Google Scholar * Wang, L. et al. Gastric mucosa-associated microbial signatures of early gastric cancer. _Front Microbiol._ 11, 1548.
https://doi.org/10.3389/fmicb.2020.01548 (2020). Article PubMed PubMed Central Google Scholar * Gunathilake, M. et al. Alterations in gastric microbial communities are associated with
risk of gastric cancer in a Korean population: A case-control study. _Cancers (Basel)._ 12(9), 2619. https://doi.org/10.3390/cancers12092619 (2020). Article CAS PubMed PubMed Central
Google Scholar * Beasley, D. E., Koltz, A. M., Lambert, J. E., Fierer, N. & Dunn, R. R. The evolution of stomach acidity and its relevance to the human microbiome. _PLoS ONE._ 10(7),
e0134116. https://doi.org/10.1371/journal.pone.0134116 (2015). Article CAS PubMed PubMed Central Google Scholar * Del Piano, M. et al. The innovative potential of Lactobacillus
rhamnosus LR06, Lactobacillus pentosus LPS01, Lactobacillus plantarum LP01, and Lactobacillus delbrueckii Subsp. delbrueckii LDD01 to restore the “gastric barrier effect” in patients
chronically treated with PPI: A pilot study. _J Clin Gastroenterol._ 46(Suppl), S18-26. https://doi.org/10.1097/MCG.0b013e318267b55d (2012). Article PubMed Google Scholar * Gray, J. D.
& Shiner, M. Influence of gastric pH on gastric and jejunal flora. _Gut._ 8(6), 574–581. https://doi.org/10.1136/gut.8.6.574 (1967). Article CAS PubMed Google Scholar *
Azcarate-Peril, M. A., Altermann, E., Hoover-Fitzula, R. L., Cano, R. J. & Klaenhammer, T. R. Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid
tolerance. _Appl Environ Microbiol._ 70, 5315–5322. https://doi.org/10.1128/AEM.70.9.5315-5322.2004 (2004). Article ADS CAS PubMed PubMed Central Google Scholar * Shin, C. M. et al.
Impact of long-term proton pump inhibitor therapy on gut microbiota in F344 rats: Pilot study. _Gut Liver._ 10, 896–901 (2016). Article CAS PubMed PubMed Central Google Scholar *
Hussain, S. P., Hofseth, L. J. & Harris, C. C. Radical causes of cancer. _Nat. Rev. Cancer._ 3, 276–285. https://doi.org/10.1038/nrc1046 (2003). Article CAS PubMed Google Scholar *
Roșca, M. F. et al. Leuconostoc citreum: A promising sourdough fermenting starter for low-sugar-content baked goods. _Foods._ 13(1), 96. https://doi.org/10.3390/foods13010096 (2023). Article
MathSciNet CAS PubMed PubMed Central Google Scholar * Cuervo, M. S. I., Cortés, L. J., Rodríguez, R. E., Hormaza, A. N. & Vargas, S. E. Leuconostoc sp en pacientes con cáncer:
Estudio descriptivo [Leuconostoc sp in cancer patients: A descriptive study]. _Rev Chilena Infectol._ 25(3), 184–8 (2008). Article Google Scholar * Dhodapkar, K. M. & Henry, N. K.
Leuconostoc bacteriemia in an infant with short-gut syndrome: Case report and literature review. _Mayo Clin Proc._ 71, 1171–1174. https://doi.org/10.4065/71.12.1171 (1996). Article CAS
PubMed Google Scholar * Espinoza, R. et al. Leuconostoc bacteremia after liver transplantation: Another cause of vancomycin resistant gram-positive infection. _Clin Transp._ 11, 322–324
(1997). Article CAS Google Scholar * Jofré, L. et al. Infección por Leuconostoc en pacientes con síndrome de intestino corto, nutrición parenteral y alimentación enteral continua. _Rev
Chil Infect._ 23, 340–345 (2006). Article Google Scholar * Fauchais, A. L. et al. Rare opportunistic infection due to Leuconostoc. _Rev Med Interne._ 24, 268–75.
https://doi.org/10.1016/s0248-8663(03)00032-8 (2003). Article PubMed Google Scholar * Nouri, Z. et al. Exploring connections between oral microbiota, short-chain fatty acids, and specific
cancer types: A study of oral cancer, head and neck cancer, pancreatic cancer, and gastric cancer. _Cancers (Basel)._ 15(11), 2898. https://doi.org/10.3390/cancers15112898 (2023). Article
CAS PubMed PubMed Central Google Scholar * Sękowska, A. Raoultella spp.-clinical significance, infections and susceptibility to antibiotics. _Folia Microbiol._ 62, 221–7.
https://doi.org/10.1007/s12223-016-0490-7 (2017). Article CAS Google Scholar * Appel, T. M., Quijano-Martínez, N., De La Cadena, E., Mojica, M. F. & Villegas, M. V. Microbiological
and clinical aspects of Raoultella spp. _Front Public Health._ 9, 686789. https://doi.org/10.3389/fpubh.2021.686789 (2021). Article PubMed PubMed Central Google Scholar * Yamamoto, S.,
Nagatani, K., Sato, T., Ajima, T. & Minota, S. Raoultella planticola bacteremia in a patient with early gastric cancer. _Intern Med._ 57(10), 1469–1473.
https://doi.org/10.2169/internalmedicine.9611-17 (2018). Article PubMed Google Scholar * Makovska, M. et al. Species and strain variability among Sarcina isolates from diverse mammalian
hosts. _Animals (Basel)._ 13(9), 1529. https://doi.org/10.3390/ani13091529 (2023). Article PubMed PubMed Central Google Scholar * Dumitru, A. et al. Fatal outcome of gastric perforation
due to infection with Sarcina spp. A case report. _IDCases._ 19, e00711. https://doi.org/10.1016/j.idcr.2020.e00711 (2020). Article PubMed PubMed Central Google Scholar * Singh, K.
Emphysematous gastritis associated with Sarcina ventriculi. _Case Rep. Gastroenterol._ 13, 207–213. https://doi.org/10.1159/000499446 (2019). Article PubMed PubMed Central Google Scholar
* de Meij, T. G., van Wijk, M. P., Mookhoek, A. & Budding, A. E. Ulcerative gastritis and esophagitis in two children with Sarcina ventriculi infection. _Front. Med._ 4, 145.
https://doi.org/10.3389/fmed.2017.00145 (2017). Article Google Scholar * Tartaglia, D. et al. Sarcina Ventriculi infection: A rare but fearsome event. A systematic review of the
literature. _Int. J. Infect. Dis._ 115, 48–61. https://doi.org/10.1016/j.ijid.2021.11.027 (2022). Article PubMed Google Scholar * Haroon Al Rasheed, M. R., Kim, G. J. & Senseng, C. A
rare case of Sarcina ventriculi of the stomach in an asymptomatic patient. _Int. J. Surg. Pathol._ 24(2), 142–145. https://doi.org/10.1177/1066896915610196 (2016). Article PubMed Google
Scholar * Sauter, J. L. et al. Co-existence of Sarcina organisms and helicobacter pylori gastritis/duodenitis in pediatric siblings. _J Clin Anat Pathol._ 1(1), 103.
https://doi.org/10.17303/jcap.2013.103 (2013). Article Google Scholar * Ugarte Bilbao, A. et al. Sarcina ventriculi in gastric biopsies of two patients with an underlying neoplasia. _Rev
Esp Enferm Dig._ 114(9), 557. https://doi.org/10.17235/reed.2022.8667/2022 (2022). Article PubMed Google Scholar * Rams, T. E. et al. Centipeda periodontii in human periodontitis.
_Odontology._ 103(3), 286–291. https://doi.org/10.1007/s10266-014-0166-1 (2015). Article CAS PubMed Google Scholar * Celik, Z. C. et al. The complex microbiome of caries-active and
caries-free supragingival plaques in permanent dentition. _Niger J Clin Pract._ 24(10), 1535–1540. https://doi.org/10.4103/njcp.njcp_49_21 (2021). Article CAS PubMed Google Scholar *
Wang, Y. et al. Alterations in the oral and gut microbiome of colorectal cancer patients and association with host clinical factors. _Int J Cancer_ https://doi.org/10.1002/ijc.33596 (2021).
Article PubMed PubMed Central Google Scholar * Granato, D. C. et al. Meta-omics analysis indicates the saliva microbiome and its proteins associated with the prognosis of oral cancer
patients. _Biochim Biophys Acta Proteins Proteom._ 1869(8), 140659. https://doi.org/10.1016/j.bbapap.2021.140659 (2021). Article CAS PubMed Google Scholar * Plotnikova, E. G., Anan’ina,
L. N., Ariskina, E. V. & Evtushenko, L. I. Salinicola. _Bergey’s Manual of Systematics of Archaea and Bacteria_ https://doi.org/10.1002/9781118960608.gbm01719 (2024). Article Google
Scholar Download references ACKNOWLEDGEMENTS The authors would like to thank the Genomics and Bioinformatics Center (CeGenBio) of Drug Research and Development Center of Federal University
of Ceara for technical support. FUNDING This work was supported by Fundação Cearense de Apoio ao Desenvolvimento Científico e Tecnológico (FUNCAP), process no. 07939716/2020; SHBR and VMMM
thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for their fellowship of research. AUTHOR INFORMATION Author notes * Silvia Helena Barem Rabenhorst and Adriana
Camargo Ferrasi contributed equally to this work. AUTHORS AND AFFILIATIONS * Genetic Molecular Laboratory, Pathology and Forensic Medicine Department, Federal University of Ceará, Fortaleza,
CE, Brazil Silvia Helena Barem Rabenhorst & Morgana Maria de Oliveira Barboza * Department of Internal Medicine, Botucatu Medical School, Sao Paulo State University, Botucatu, Brazil
Adriana Camargo Ferrasi * Microbial Ecology and Biotechnology Laboratory, Department of Biology, Federal University of Ceará, Fortaleza, CE, Brazil Vânia Maria Maciel Melo Authors * Silvia
Helena Barem Rabenhorst View author publications You can also search for this author inPubMed Google Scholar * Adriana Camargo Ferrasi View author publications You can also search for this
author inPubMed Google Scholar * Morgana Maria de Oliveira Barboza View author publications You can also search for this author inPubMed Google Scholar * Vânia Maria Maciel Melo View author
publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Conceptualization: SHBR, ACF, VMMM; Data curation: SHBR, ACF, VMMM, MMOB; Project administration: SHBR;
Manuscript draft: SHBR, ACF; Review and editing: SHBR, ACF, VMMM; Approval of final version: all.**SHBR and ACF have contributed equally to this work and share first authorship.
CORRESPONDING AUTHOR Correspondence to Adriana Camargo Ferrasi. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ETHICS APPROVAL The study was approved by
Ethics Committee (COMEP) of the Federal University of Ceará (protocol 071002/10) and conducted in accordance with the Declaration of Helsinki. All participants of the study signed individual
informed consent forms. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. RIGHTS
AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use,
sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons
licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or
other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in
the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the
copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Rabenhorst, S.H.B.,
Ferrasi, A.C., Barboza, M.d. _et al._ Microbial composition of gastric lesions: differences based on _Helicobacter pylori_ virulence profile. _Sci Rep_ 14, 28890 (2024).
https://doi.org/10.1038/s41598-024-80394-2 Download citation * Received: 27 July 2024 * Accepted: 18 November 2024 * Published: 21 November 2024 * DOI:
https://doi.org/10.1038/s41598-024-80394-2 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative KEYWORDS * Helicobacter pylori * Gastric cancer * Metagenomics