Play all audios:
ABSTRACT Head and neck squamous cell carcinoma (HNSCC) is a heterogeneous cancer with limited therapeutic options. Using publicly available datasets, we identified the WD repeat domain 54
(_WDR54_) gene as a potential therapeutic target in HNSCC. Gene expression profiling interactive analysis version 2 (GEPIA2) was used to identify genes differentially overexpressed in HNSCC.
Our results showed that WDR54, a member of the WD40 repeat domain family, was overexpressed in HNSCC tumor samples. Analysis of three gene expression omnibus datasets showed that WDR54 was
overexpressed in tumor samples. Using the UALCAN database, we showed that WDR54 expression in patients with HNSCC at different tumor stages gradually increased with disease progression. We
confirmed the association between WDR54 and metastasis using TNMplot.com. WDR54 was overexpressed in metastatic samples compared to that in normal and tumor samples. Kaplan–Meier analysis
showed that patients with high WDR54 levels had a poorer prognosis. Additionally, WDR54 expression was correlated with the epidermal growth factor receptor, which is frequently overexpressed
in HNSCC. Our findings suggest that WDR54 is a promising biomarker and therapeutic target in HNSCC. SIMILAR CONTENT BEING VIEWED BY OTHERS DEVELOPMENT AND VALIDATION OF A PREDICTIVE MODEL
BASED ON Β-KLOTHO FOR HEAD AND NECK SQUAMOUS CELL CARCINOMA Article Open access 24 July 2024 DATA MINING ON IDENTIFYING DIAGNOSIS AND PROGNOSIS BIOMARKERS IN HEAD AND NECK SQUAMOUS CARCINOMA
Article Open access 20 June 2023 MULTI-OMICS IN IMMUNOTHERAPY RESEARCH FOR HNSCC: PRESENT SITUATION AND FUTURE PERSPECTIVES Article Open access 29 March 2025 INTRODUCTION Head and neck
squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide1. The key factors contributing to HNSCC carcinogenesis include tobacco use and human papillomavirus (HPV)
infection2. Cornerstone treatments for HNSCC include surgery, radiotherapy, and chemotherapy. However, these interventions often result in significant morbidity and do not sufficiently
address the high rates of metastasis and recurrence. Despite advances in therapeutic techniques, the estimated 5-year survival rate of patients with HNSCC remains below 50%3. This stagnation
in therapeutic advancements highlights the urgent need for novel diagnostic and therapeutic strategies for the management of HNSCC. Biomarkers have revolutionized oncological diagnostics
and therapies, creating new opportunities for personalized treatment regimens. In HNSCC, as in other malignancies, identifying accurate biomarkers is critical for the early detection,
prediction of disease progression, and response to therapy4. However, the discovery of effective tailored treatments for HNSCC has been impeded by a lack of knowledge regarding the molecular
mechanisms that promote tumor growth and resistance to treatment. Recent advancements in bioinformatics have revolutionized cancer research by providing tools that can systematically
analyze vast datasets to uncover potential genetic drivers of cancer. This approach is particularly promising for the identification of novel therapeutic targets specific to the genetic
landscape of individual cancers. The gene expression profiling interactive analysis version 2 (GEPIA2) is a web server that provides robust gene expression profiling and is instrumental in
advancing our understanding of cancer biology5. Our study leveraged the capabilities of GEPIA2 to investigate WD repeat domain 54 (WDR54), which has previously been underexplored in the
context of HNSCC. The WDR domain is composed of a seven-bladed ß-propeller structure that ends with a tryptophan-aspartic acid dipeptide composed of 40 amino acids. The WDR domain is one of
the most abundant protein-protein interaction (PPI) domains6. PPIs play various roles in cell cycle regulation7,8, signal transduction9, and apoptosis10. Several WDR domains have been
implicated in carcinogenesis11,12,13 and metastasis in various cancers, such as gastric, lung, and prostate cancers. With the expanding understanding of PPIs, the human protein interactome
is increasingly being recognized as a rich source of potential targets for disease treatment14,15. WDR54 plays an oncogenic role in bladder and colon cancers and T-cell acute lymphoblastic
leukemia16,17,18. Although WDR domains have been studied in various cancer types, their roles in HSNCC have not yet been investigated. We conducted a comprehensive analysis of the biological
functions of WDR54 in HNSCC using GEPIA2 and the gene expression omnibus (GEO) database. We observed that WDR54 was overexpressed in HNSCC and associated with poor patient survival. These
findings suggest that WDR54 is a potential prognostic biomarker and therapeutic target for patients with HNSCC. The purpose of this study was to fully characterize the expression of WDR54 in
HNSCC using bioinformatic methods and to assess its potential as a therapeutic target. By defining the role of WDR54 in the pathophysiology of HNSCC and conducting additional in vitro
experiments, we expect to contribute to the development of more precise and successful treatment options, thereby improving patient outcomes for this challenging disease. RESULTS WDR54
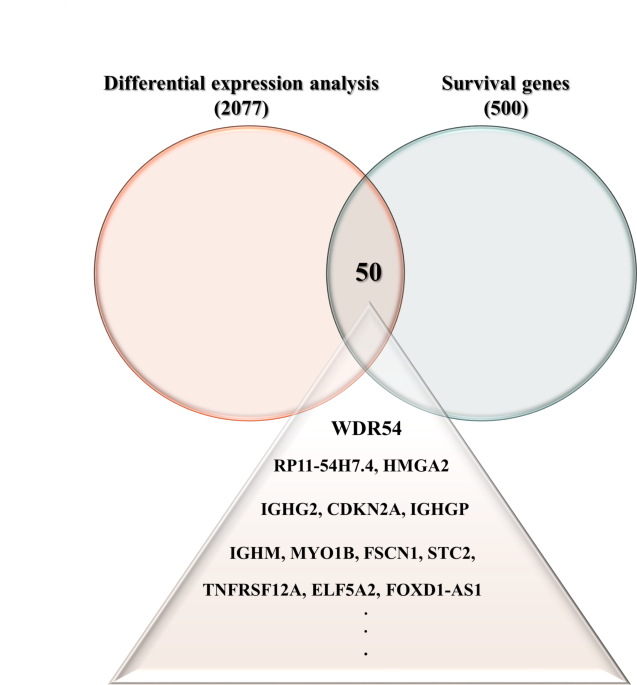
EXPRESSION IS UPREGULATED IN HNSCC We used GEPIA2 to search for genes that were commonly expressed as part of the survival-related gene category (_n_ = 500) and genes that were
differentially expressed (_n_ = 2077) between normal and HNSCC tumor tissues using the prostate adenocarcinoma (TCGA-PRAD) and Genotype-Tissue Expression (GTEx) datasets accessed from the
Cancer Genome Atlas. We identified 50 genes that were commonly overexpressed in both differential expression and survival analyses of HNSCC. We selected WDR54 and analyzed the effect on
HNSCC (Fig. 1). Using the GEPIA2 database, we compared 44 normal and 519 tumor samples. WDR54 was overexpressed in most tumors compared to that in normal tissues, particularly lymphoid
neoplasm diffuse large B cell lymphoma (DLBC), esophageal carcinoma (ESCA), kidney chromophobe (KIRC), kidney renal clear cell carcinoma (KIRP), pancreatic adenocarcinoma (PAAD), skin
cutaneous melanoma (SKCM), thymoma (THYM), and head and neck squamous cell carcinoma (HNSC) (Fig. 2A). A comparison of WDR54 mRNA expression in 44 normal and 519 HNSCC tumor tissues revealed
significant overexpression in tumor tissues (Fig. 2B). Especially, the expression level of WDR54 was significantly higher in the HNSCC tumors compared with matched normal tissues (Fig. 2C).
We used the GSE58911, GSE178537, and GSE33232 datasets to analyze WDR54 expression and observed that it was significantly elevated in tumors compared to that in normal samples in all three
analyses (Fig. 2D). WDR54 LEVELS AND CLINICAL CHARACTERISTICS OF PATIENTS WITH HNSCC We have identified that WDR54 is overexpressed in HNSCC and analyzed whether there is a difference in
WDR54 expression among different pathological stages using the online UALCAN database. WDR54 levels were observed to increase with cancer stage (stages 1, 2, 3, and 4) in patients with HNSCC
compared to those in normal tissues (Fig. 3A). WDR54 expression was higher in HNSCC tissues classified as cancer stages 1, 2, 3, and 4 than that in normal tissues (Fig. 3B). In the case of
nodal metastasis, the expression of WDR54 at all stages (N0, N1, N2, and N3) was higher than that in normal tissues (Fig. 3C). WDR54 EXPRESSION IN HNSCC METASTASIS We performed an
RNA-seq-based TNM plot analysis to compare normal samples with metastatic patient samples to predict whether WDR54 could influence metastasis. We observed that WDR54 expression was further
increased in metastatic patient samples compared to that in normal samples (Fig. 4A). Additionally, we analyzed WDR54 expression using several GEO datasets (GSE178537, GSE117753, and
GSE67614) and observed that metastatic samples had significantly higher WDR54 levels than that in normal and tumor samples (Fig. 4B). PROGNOSTIC VALUE OF WDR54 LEVELS IN HNSCC To further
clarify the relationship between WDR54 expression and the prognosis of patients with HNSCC, we used the GEPIA2 database and Kaplan–Meier survival curve analysis. Using a survival map from
the GEPIA2 database, we analyzed the expression of genes in various cancer types and observed that increased WDR54 expression was associated with poor survival in patients with
adrenocortical carcinoma, acute myeloid leukemia, liver hepatocellular carcinoma, mesothelioma, uveal melanoma, and HNSCC (Fig. 5A). An overall survival graph obtained using the GEPIA2
database confirmed that WDR54 overexpression was associated with poor survival (_p_ = 0.00023, hazard ratio [HR] (high) = 2.1, p (HR) = 0.00031) (Fig. 5B). Similar to the GEPIA2 database,
the Kaplan–Meier plotter analysis showed that patients with high WDR54 expression had lower survival rates, with a median overall survival of 61.27 months for WDR54 low expression and 32.83
months for WDR54 high expression (_p_ = 0.0031, HR = 1.49) (Fig. 5C). We analyzed the survival rates according to tumor stage and WDR54 levels. When we checked by pathological stage, we
observed that a high WDR54 level was associated with poor survival in stages 1 + 2, 3, and 4 (Fig. 5D and F). Although the results were not significant when stages 1 and 2 were examined
separately, high WDR54 levels were associated with poor survival (data not shown). Patients with high WDR54 levels had lower survival rates than those with low WDR54 levels at all stages.
The survival rates decreased gradually with high WDR54 levels at various tumor stages, progressing from 85.67 months for stage 1 + 2 (_p_ = 0.059, HR = 2.67), to 32.83 months for stage 3
(_p_ = 0.046, HR = 2.37), and finally declining to 30.5 months for stage 4 (_p_ = 0.026, HR = 1.52). When analyzed by sex, both men and women with higher WDR54 levels had lower survival
rates (Supplementary Fig. S1A, S1B). Survival rates were 35.97 months for men and 25.43 months for women with high WDR54 levels. We observed a significant correlation between WDR54
expression and the immune subtype in HNSCC. We performed a Kaplan–Meier survival analysis by stratifying the active immune subtypes. Patients with high WDR54 levels had a poor prognosis,
regardless of whether they had enriched or decreased levels of macrophages, natural killer T cells, CD4 + memory T cells, or CD8 + T cells (Fig. 6 and Supplementary Fig. S2). CORRELATION
BETWEEN WDR54 AND EPIDERMAL GROWTH FACTOR RECEPTOR (EGFR) EXPRESSION We used the GEPIA2 online tool to analyze the correlation between the human EGFR and WDR54 proteins, which are target
biomarkers for head and neck cancer and various other cancers. We observed that both genes were overexpressed in tumors compared to those in normal tissues in the DLBS, ESCA, GBM, KIRC,
KIRP, LGG, LUSC, PAAD, STAD, THYM, and HNSCC gene expression datasets. (Fig. 7A). Moreover, WDR54 expression was positively correlated with EGFR expression in HNSCC (Fig. 7B). We further
examined the expression of WDR54 and EGFR using two different GEO datasets (GSE58911 and GSE178537). Both datasets indicated that WDR54 expression was significantly correlated with EGFR
expression (Fig. 7C). ANALYZING THE THERAPEUTIC TARGET POTENTIAL OF WDR54 IN HNSCC CELL To assess the therapeutic target potential of WDR54 in HNSCC, we employed siRNA to reduce the
expression of WDR54 in FaDu cell (Fig. 8A). WDR54-downregulated FaDu cell decreased proliferation (Fig. 8B). We treated WDR54-downregulated FaDu cell with varying concentrations of the
commonly used chemotherapeutic agents for HNSCC, cisplatin and 5-fluorouracil, to assess their cell viability. The results showed that WDR54- downregulated FaDu cell exhibited increased
sensitivity to cisplatin and 5-fluorouracil (Fig. 8C–D). This suggests that WDR54 may serve as a novel therapeutic target for HNSCC. DISCUSSION HNSCC is a highly heterogeneous cancer
characterized by an aggressive disease course, a high recurrence rate, a low response to treatment, and a low survival rate, leading to a need for research on its pathogenesis and new
therapeutic agents. We accessed the publicly available GEPIA2 web server to identify differentially overexpressed genes in HNSCC and analyzed various GEO datasets to identify novel
therapeutic targets for head and neck cancer. We screened several candidate genes using GEPIA2 and selected WDR54 for further analysis. WDR54 is a member of the WD40 repeat-domain family.
WDR is a PPI domain19,20 that plays diverse roles in cell cycle regulation7,8, signal transduction9, and apoptosis10, and several WDR domains have been implicated in carcinogenesis11,12,13.
In the current study, we examined the expression of WDR54, a post-translationally modified WDR domain protein21. WDR54 plays an oncogenic role in various cancers, and our data support its
potential as a novel therapeutic target and biomarker for head and neck cancer, based on our analysis of the GEPIA2, GEO, and UALCAN gene expression datasets16,17,18. We observed that WDR54
expression was significantly higher in patients with HNSCC than that in healthy individuals. Using TNM plot, a web tool for normal, tumor, and metastatic tissue gene expression, we compared
tumor and metastatic samples to show that WDR54 was overexpressed in metastatic samples compared to that in tumor samples. We initially observed an increase in WDR54 expression in tumor and
metastasis samples compared to normal samples using TNMplot analysis, but the analysis was limited due to only two metastasis samples. To address this limitation, we conducted further
analysis using the GSE178537, GSE117753, and GSE67614 datasets, which include normal, tumor, and metastasis samples. Our results confirmed that WDR54 was significantly overexpressed in
metastasis samples, providing additional support for the findings obtained from the TNMplot analysis. Together, these findings suggest that WDR54 overexpression contributes to cancer
progression and metastasis in HNSCC cells. We used GEPIA2 and Kaplan–Meier analyses to determine whether high WDR54 expression was associated with HNSCC survival. Our overall survival
analysis using both GEPIA2 and Kaplan–Meier analyses showed that high WDR54 expression levels were associated with poor survival outcomes in patients with HNSCC, regardless of sex, disease
stage, immune subtype enrichment, or depletion. These findings suggest that WDR54 is a potential novel therapeutic target for HNSCC. _EGFR_ is an oncogene that affects gene expression,
proliferation, angiogenesis, apoptosis inhibition, cell motility, metastasis, adhesion, and angiogenesis. HNSCC is a highly heterogeneous disease characterized by the overexpression of EGFR.
Over 90% of the patients with HNSCC overexpressing EGFR have a shorter survival period, which is one of the reasons why cetuximab, which targets EGFR, is used to treat HNSCC22. WDR54 has
been shown to be cross-linked by transglutaminase 2, and this process has been found to inhibit the activity of EGFR-mediated signaling tumorigenesis23. We identified a correlation between
WDR54 and the overexpression of EGFR in head and neck cancer. This association suggests that WDR54 may play a role in modulating EGFR-driven oncogenic processes. WDR54 has been identified as
a novel oncogene in colorectal18 and bladder cancer16 and acute leukemia17, and it is known to be associated with cell growth. We used FaDu cells, which are commonly used to evaluate the
efficacy of radiotherapy and chemotherapy, to determine the impact of WDR54 in HNSCC. We found that FaDu cells with reduced WDR54 exhibited decreased cell proliferation, similar to other
cancers, and showed significantly increased sensitivity to the head and neck cancer drugs cisplatin and 5-fluorouracil. This suggests that targeting WDR54 could potentially improve the
efficacy of these chemotherapeutic agents, offering a promising avenue for therapeutic intervention in HNSCC. CONCLUSION We observed a positive correlation between EGFR and WDR54 expression
in HNSCC cells. Our analysis using only tumor samples showed a robust correlation between EGFR and WDR54, suggesting that WDR54 may act as an oncogene and regulator of EGFR expression in
HNSCC. Previous studies have reported the role of WDR54 in several cancers; however, its function in HNSCC remains poorly understood. Based on a comprehensive analysis of publicly available
gene expression datasets, we propose WDR54 as a potential new biomarker and therapeutic target for HNSCC. However, further experimental studies are required to elucidate the function and
molecular mechanism of action of WDR54 in HNSCC. Such studies will provide a better understanding of the role of WDR54 in HNSCC and lead to the development of novel therapeutic strategies
for this disease. MATERIALS AND METHODS GENE EXPRESSION PROFILING INTERACTIVE ANALYSIS VERSION 2 (GEPIA2) To quantitatively assess WDR54 expression in various tissues and its association
with clinical outcomes in HNSCC, we used the GEPIA2 platform, an advanced online resource designed for comprehensive analysis of RNA sequencing data. Hosted at “gepia2.cancer-pku.cn,” this
tool facilitates robust and detailed investigations of gene expression patterns by integrating data from two major databases: The Cancer Genome Atlas (TCGA) and the Genotype-Tissue
Expression (GTEx) project5,24. MICROARRAY DATA The National Center for Biotechnology Information (NCBI) GEO database (http://www.ncbi.nlm.nih.gov/geo/) is a publicly available resource
containing a wealth of high-throughput functional genomic data. The data used in our analysis were obtained from the NCBI-GEO database by searching for the keywords “Head and neck squamous
cell carcinoma” and “homo sapiens,” and the datasets GSE 33,232, GSE 58,911, GSE 178,537, GSE 117,753, and GSE 67,614 were used for analysis (Table 1). The clinical characteristics and
detailed information are summarized in Supplementary Tables 1 and 2, respectively. KAPLAN–MEIER PLOTTER ANALYSIS The Kaplan–Meier plotter (http://kmplot.com/analysis/) enables the assessment
of the relationship between the expression of all genes (mRNA, miRNA, Protein, and DNA) and tumor survival using data from > 35,000 samples across 21 different types of
cancer25,26,27,28,29,30. The correlation between WDR54 expression and clinical characteristics in Kaplan-Meier plotter are summarized in Supplementary Table 3. UALCAN UALCAN
(http://ualcan.path.uab.edu/) is a publicly accessible web-based platform that enables the analysis of cancer omics data, with a particular focus on tumor gene expression and survival
analysis31. The clinical information of HNSC patients in UALCAN database are summarized in Supplementary table 4A–D. TNMPLOT TNM plot (http://www.tnmplot.com/) is an online tool that
conducts differential gene expression analysis of tumor, normal, and metastatic tissues. Moreover, it utilizes data from 56,938 unique samples, including 15,648 normal, 40,442 tumors, and
848 metastatic samples collected from the GEO, GTEx, TCGA, and TARGET databases32. The detailed clinical characteristics in TNM plot database are summarized in Supplementary table 5A and 5B.
SMALL INTERING RNA(SIRNA) TRANSFECTION FaDu cell was transfected with siRNA for WDR54 and negative control (NC) (Bioneer, Daejeon, Korea) using the RNAiMAX transfection reagent (Thermo
Fisher Scientific) according to the manufacturer’s instructions. The siRNA sequences used are listed in Table 2. TOTAL RNA EXTRACTION AND QUATITATIVE REVERSE TRNASCIPTION-PCR(QRT-PCR) Total
RNA was extracted using Nucleozol (MACHEREY-NAGEL GmbH & Co., KG, Düren, Germany) according to the manufacturer’s instructions. cDNA was synthesised with PrimeScript™ RT reagent Kit
(Takara, Kusatsu, Shiga, Japan), according to the manufacturer‘s instructions. Quantitative real-time PCR was performed by a Step One Plus real-time PCR system (Applied Biosystems, Waltham,
MA, USA) using Fast SYBR Green Master Mix (Applied Biosystems). β-ACTIN mRNA expression levels were used to normalise mRNA expression levels. Primer sequences used in this study are listed
in Table 3. CELL CULTURE AND ANALYSIS OF CELL VIABILITY FaDu cell was obtained from the American Type Culture Collection (ATCC) and were grown in Minimum Essential Medium (MEM) supplemented
with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, and 1% sodium pyruvate. The cells were maintained at 37 °C in a humidified incubator with 5% CO2. Cell viability and cell
proliferation was measured using an EZ-Cytox Cell Viability Assay Kit (Dogenbio, Seoul, Korea). Cell viability and proliferation were analyzed using WDR54-knockdown FaDu cell. For cell
viability, 3 × 103 cells were seeded per well in a 96-well plate, and for cell proliferation, 1 × 103 cells were seeded per well. Cisplatin and 5-Fluorouracil (Sigma-Aldrich, St. Louis, MO,
USA) were added to each well and incubated for 72 h with FaDu cell. Then, 10 µL EZ-Cytox was added into the well and measured at 450 nm using a microplate reader (Bio–Tek, Winooski, VT,
USA). STATISTICAL ANALYSIS The data was analyzed using GraphPad Prism 8.0 software, and statistical tests included ANOVA, Tukey’s multiple comparison test, and Student’s t-test. All data is
presented as the mean ± SD, and statistical significance was set at _p_ < 0.05. DATA AVAILABILITY The data that support the findings of this study can be made available from the
corresponding author, Y.S.Kim, upon reasonable request. REFERENCES * Gormley, M., Creaney, G., Schache, A. & Ingarfield, K. Conway, D. I. reviewing the epidemiology of head and neck
cancer: Definitions, trends and risk factors. _Br. Dent. J._ 233, 780–786 (2022). Article PubMed PubMed Central Google Scholar * Katiyar, S. K. Emerging phytochemicals for the prevention
and treatment of head and neck cancer. _Molecules_ 21, 1610 (2016). Article PubMed PubMed Central MATH Google Scholar * de Andrade, D. A. & Machiels, J. P. Treatment options for
patients with recurrent or metastatic squamous cell carcinoma of the head and neck, who progress after platinum-based chemotherapy. _Curr. Opin. Oncol._ 24, 211–217 (2012). Article PubMed
MATH Google Scholar * Johnson, D. E. et al. Head and neck squamous cell carcinoma. _Nat. Rev. Dis. Primers_. 6, 92 (2020). Article PubMed PubMed Central MATH Google Scholar * Tang,
Z., Kang, B., Li, C., Chen, T., Zhang, Z. & GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. _Nucleic Acids Res._ 47, W556–W560 (2019).
Article PubMed PubMed Central Google Scholar * Schapira, M., Tyers, M., Torrent, M. & Arrowsmith, C. H. WD40 repeat domain proteins: A novel target class? _Nat. Rev. Drug Discov_.
16, 773–786 (2017). Article PubMed PubMed Central Google Scholar * An, H. J. et al. FBXW7-mediated ERK3 degradation regulates the proliferation of lung cancer cells. _Exp. Mol. Med._ 54,
35–46 (2022). Article PubMed PubMed Central MATH Google Scholar * Jain, B. P. & Pandey, S. WD40 repeat proteins: Signalling scaffold with diverse functions. _Protein J._ 37,
391–406 (2018). Article PubMed MATH Google Scholar * Sun, Z., Tang, X., Lin, F. & Chen, S. The WD40 repeat protein WDR26 binds Gbetagamma and promotes gbetagamma-dependent signal
transduction and leukocyte migration. _J. Biol. Chem._ 286, 43902–43912 (2011). Article PubMed PubMed Central Google Scholar * Saeki, M. et al. Monad, a WD40 repeat protein, promotes
apoptosis induced by TNF-alpha. _Biochem. Biophys. Res. Commun._ 342, 568–572 (2006). Article PubMed Google Scholar * Kim, J. Y. et al. A role for WDR5 in integrating threonine 11
phosphorylation to lysine 4 methylation on histone H3 during androgen signaling and in prostate cancer. _Mol. Cell._ 54, 613–625 (2014). Article PubMed PubMed Central Google Scholar *
Zuo, J., Liu, C., Ni, H. & Yu, Z. WDR34 affects PI3K/Akt and Wnt/beta-catenin pathways to regulates malignant biological behaviors of glioma cells. _J. Neurooncol_. 156, 281–293 (2022).
Article PubMed Google Scholar * Zhang, Y. et al. Overexpression of WDR62 is associated with centrosome amplification in human ovarian cancer. _J. Ovarian Res._ 6, 55–60 (2013). Article
ADS PubMed PubMed Central Google Scholar * Huttlin, E. L. et al. The BioPlex network: a systematic exploration of the human interactome. _Cell_ 162, 425–440 (2015). Article PubMed
PubMed Central Google Scholar * Huttlin, E. L. et al. Architecture of the human interactome defines protein communities and disease networks. _Nature_ 545, 505–509 (2017). Article ADS
PubMed PubMed Central Google Scholar * Wei, X. et al. WD repeat protein 54-mediator of ErbB2-driven cell motility 1 axis promotes bladder cancer tumorigenesis and metastasis and impairs
chemosensitivity. _Cancer Lett._ 556, 216058 (2023). Article PubMed Google Scholar * Li, H. et al. WDR54 exerts oncogenic roles in T-cell acute lymphoblastic leukemia. _Cancer Sci._ 114,
3318–3329 (2023). Article ADS PubMed PubMed Central MATH Google Scholar * Yuan, Y. et al. Clinical significance and biological function of WD repeat domain 54 as an oncogene in
colorectal cancer. _Int. J. Cancer_. 144, 1584–1595 (2019). Article PubMed MATH Google Scholar * Stirnimann, C. U., Petsalaki, E. & Russell, R. B. Muller, C. W. WD40 proteins propel
cellular networks. _Trends Biochem. Sci._ 35, 565–574 (2010). Article PubMed Google Scholar * Xu, C. & Min, J. Structure and function of WD40 domain proteins. _Protein Cell._ 2,
202–214 (2011). Article PubMed PubMed Central MATH Google Scholar * Rush, J. et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. _Nat. Biotechnol._ 23, 94–101
(2005). Article PubMed MATH Google Scholar * Kalyankrishna, S. & Grandis, J. R. Epidermal growth factor receptor biology in head and neck cancer. _J. Clin. Oncol._ 24, 2666–2672
(2006). Article PubMed MATH Google Scholar * Maeda, A. et al. Transglutaminase-mediated cross-linking of WDR54 regulates EGF receptor-signaling. _Biochim. Biophys. Acta Mol. Cell. Res._
1866, 285–295 (2019). Article PubMed MATH Google Scholar * Tang, Z. et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. _Nucleic Acids
Res._ 45, W98–W102 (2017). Article PubMed PubMed Central Google Scholar * Stansfield, J. C. et al. Toward signaling-driven biomarkers immune to normal tissue contamination. Cancer
informatics. _Cancer Inf._ 15, CIN–S32468 (2016). Google Scholar * Lobert, S. et al. Prognostic biomarkers for HNSCC using quantitative real-time PCR and microarray analysis: Beta-tubulin
isotypes and the p53 interactome. _Cytoskeleton_ 71, 628–637 (2014). Article PubMed Google Scholar * Cheng, H. Y. et al. Snail-regulated exosomal microRNA-21 suppresses NLRP3 inflammasome
activity to enhance cisplatin resistance. _J. Immunother Cancer_ 10 (2022). * Kondratyev, M. et al. Identification of acquired Notch3 dependency in metastatic head and neck cancer. _Commun.
Biol._ 6, 538 (2023). Article PubMed PubMed Central MATH Google Scholar * Ding, L. et al. Evaluation of the response of HNSCC cell lines to γ-rays and 12 C ions: Can radioresistant
tumors be identified and selected for 12 C ion radiotherapy? _Front. Oncol._ 12, 812961 (2022). Article PubMed PubMed Central Google Scholar * Györffy, B. et al. An online survival
analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. _Breast Cancer Res. Treat._ 123, 725–731 (2010). Article
PubMed MATH Google Scholar * Chandrashekar, D. S. et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. _Neoplasia_ 19, 649–658 (2017). Article
PubMed PubMed Central MATH Google Scholar * Bartha, Á. & Győrffy, B. TNMplot. Com: A web tool for the comparison of gene expression in normal, tumor and metastatic tissues. _Int. J.
Mol. Sci._ 22, 2622 (2021). Article PubMed PubMed Central MATH Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by the Korea Medical Device Development Fund
grant funded by the Korea government (the Ministry of Science and ICT, Ministry of Trade, Industry and Energy, Ministry of Health & Welfare, and Ministry of Food and Drug Safety)
(Project Number: RS-2020-KD000027), and by the Global–Learning & Academic Research Institution for Master’s·PhD students, and Postdocs (LAMP) Program of the National Research Foundation
(NRF) of Korea grant funded by the Ministry of Education (Grant Number: RS-2023-00301914). FUNDING This work was supported by the Korea Medical Device Development Fund grant funded by the
Korean government (the Ministry of Science and ICT, Ministry of Trade, Industry and Energy, Ministry of Health & Welfare, and Ministry of Food and Drug Safety) (Project Number:
RS-2020-KD000027), and by the Global–Learning & Academic Research Institution for Master’s· PhD students and the Postdocs (LAMP) Program of the National Research Foundation (NRF) of
Korea grant funded by the Ministry of Education (Grant Number: RS-2023-00301914). AUTHOR INFORMATION Author notes * Eun-jeong Jeong and Eunjeong Kim are co-first authors. AUTHORS AND
AFFILIATIONS * Department of Otorhinolaryngology–Head and Neck Surgery, Konyang University College of Medicine, Daejeon, Korea Eun-jeong Jeong * Institute of Basic Sciences, BK21 FOUR KNU
Creative BioResearch Group, Department of Biology, College of Natural Sciences, KNU G-LAMP Research Center, KNU, Kyungpook National University, Daegu, 41566, Republic of Korea Eunjeong Kim *
Department of Otorhinolaryngology–Head and Neck Surgery, Korea University College of Medicine, 35365, Seoul, Korea Eun-jeong Jeong & Yeon Soo Kim Authors * Eun-jeong Jeong View author
publications You can also search for this author inPubMed Google Scholar * Eunjeong Kim View author publications You can also search for this author inPubMed Google Scholar * Yeon Soo Kim
View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Conceptulization, E.-J.J., E.J.K. and Y.S.K.; Methodology, E.-J.J. and E.J.K.; Software,
E.-J.J., and E.J.K. ; Validation, E.-J.J. and E.J.K.; Formal analysis, E.-J.J. and E.J.K.; Investigation, E.-J.J. and E.J.K.; Resources, E.-J.J. and E.J.K.; Data curation, E.-J.J. and
E.J.K.; Writing-original draft preparation, E.-J.J.; Writing-review and editing, E.J.K. and Y.S.K.; Visualization, E.-J.J. and E.J.K.; Supervision, Y.S.K.; Project administration, Y.S.K.;
Funding acquisition, Y.S.K. and E.J.K; All authors have read and agreed to the published version of the manuscript. CORRESPONDING AUTHOR Correspondence to Yeon Soo Kim. ETHICS DECLARATIONS
COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps
and institutional affiliations. ELECTRONIC SUPPLEMENTARY MATERIAL Below is the link to the electronic supplementary material. SUPPLEMENTARY MATERIAL 1. SUPPLEMENTARY MATERIAL 2.
SUPPLEMENTARY MATERIAL 3. SUPPLEMENTARY MATERIAL 4. SUPPLEMENTARY MATERIAL 5. SUPPLEMENTARY MATERIAL 6. SUPPLEMENTARY MATERIAL 7. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed
under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or
format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material.
You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the
article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use
is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit
http://creativecommons.org/licenses/by-nc-nd/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Jeong, Ej., Kim, E. & Kim, Y.S. Identification of novel therapeutic
targets for head and neck squamous cell carcinoma through bioinformatics analysis. _Sci Rep_ 14, 32102 (2024). https://doi.org/10.1038/s41598-024-83680-1 Download citation * Received: 21
June 2024 * Accepted: 16 December 2024 * Published: 30 December 2024 * DOI: https://doi.org/10.1038/s41598-024-83680-1 SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative KEYWORDS * Head and neck squamous cell carcinoma * WD repeat domain 54 * Gene expression profiling interactive analysis web server * Gene expression omnibus