Play all audios:
ABSTRACT Alzheimer’s disease (AD) is one of the primary causes of dementia in the older population, affecting memories, cognitive levels, and the ability to accomplish simple activities
gradually. Timely intervention and efficient control of the disease prove to be possible through early diagnosis. The conventional machine learning models designed for AD detection work well
only up to a certain point. They usually require a lot of labeled data and do not transfer well to new datasets. Additionally, they incur long periods of retraining. Relatively powerful
models of deep learning, however, also are very demanding in computational resources and data. In light of these, we put forward a new way of diagnosing AD using magnetic resonance imaging
(MRI) scans and transfer learned convolutional neural networks (CNN). Transfer learning makes it easier to reduce the costs involved in training and improves performance because it allows
the use of models which have been trained previously and which generalize very well even when there is very little training data available. In this research, we used three different
pre-trained CNN based architectures (AlexNet, GoogleNet, and MobileNetV2) each implemented with several solvers (e.g. Adam, Stochastic Gradient Descent or SGD, and Root Mean Square
Propagation or RMSprop). Our model achieved impressive classification results of 99.4% on the Kaggle MRI dataset as well as 98.2% on the Open Access Series of Imaging Studies (OASIS)
database. Such results serve to demonstrate how transfer learning is an effective solution to the issues related to conventional models that limits the accuracy of diagnosis of AD, thus
enabling their earlier and more accurate diagnosis. This would in turn benefit the patients by improving the treatment management and providing insights on the disease progression. SIMILAR
CONTENT BEING VIEWED BY OTHERS IDENTIFICATION OF ALZHEIMER'S DISEASE USING A CONVOLUTIONAL NEURAL NETWORK MODEL BASED ON T1-WEIGHTED MAGNETIC RESONANCE IMAGING Article Open access 17
December 2020 THREE-ROUND LEARNING STRATEGY BASED ON 3D DEEP CONVOLUTIONAL GANS FOR ALZHEIMER’S DISEASE STAGING Article Open access 07 April 2023 STRUCTURE FOCUSED NEURODEGENERATION
CONVOLUTIONAL NEURAL NETWORK FOR MODELLING AND CLASSIFICATION OF ALZHEIMER’S DISEASE Article Open access 03 July 2024 INTRODUCTION Alzheimer’s disease (AD) is a type of dementia that
primarily affects older adults, causing significant brain tissue damage and the death of nerve cells. This leads to memory loss and difficulties with daily activities such as reading,
writing, speaking, and recognizing family members1. In advanced stages, patients may struggle to recognize loved ones and experience severe symptoms like heart failure and respiratory
dysfunction, which can be fatal2. The symptoms of AD gradually worsen over time, severely impacting overall health3. By 2050, it is estimated that one in every 85 individuals will suffer
from AD, underscoring its increasing prevalence4. Around 60 to 80% of AD patients exhibit dementia symptoms, making it the world’s second most severe neurological disorder5. The disease
causes the cerebral cortex and hippocampus to shrink while the brain’s ventricles enlarge. The reduction in hippocampal size impairs episodic and spatial memory, leading to problems with
judgment, short-term memory, and planning6. Machine learning and deep learning algorithms have proven highly effective in diagnosing a wide range of diseases, including those affecting the
heart, lungs, brain, retina, breast, and bones7,8,9,10,11,12. These advanced methods play a vital role in medical image classification, a key area of computational intelligence13,14,15,16.
Convolutional neural networks (CNNs) stand out in deep learning due to their exceptional accuracy in medical image classification. A significant advantage of CNNs over traditional machine
learning techniques is their ability to automatically extract the most relevant features and classify the stages of Alzheimer’s disease, eliminating the need for manual feature extraction17.
Nevertheless, several such works still reported accuracy, generalizability, and data efficiency issues when using MRI data for Alzheimer’s classification. The high dimensionality of MRI
data makeup makes the features interdependent and hence there is needed for models which can extract useful features with very little manual effort. There are several studies which utilized
MRI data and different AI techniques for the classification glioma, meningioma, and pituitary tumors18,19,20,21 Several recent studies have examined the reliability of MRI for diagnosing
Alzheimer’s disease. For instance, Zhang et al.22 proposed a hybrid machine learning approach that requires retraining when new data is introduced, which limits its scalability in clinical
applications. Similarly, Liu et al.23 explored the use of auto-encoders for classifying Alzheimer’s stages. While the results were promising, the method required significant data
preprocessing, which can be resource intensive. Additionally, Bilfaqih et al.24 developed a hybrid deep learning approach that achieved 98.5% accuracy using a combination of MRI and other
imaging modalities, but the method was constrained by its binary classification output, limiting its broader applicability in distinguishing between multiple stages of Alzheimer’s disease.
Our study builds upon the advancements of those prior works in that we are able to diagnose fast and accurately thanks to transfer learning using pre-trained CNNs even for datasets that are
relatively small for Alzheimer’s recognition. In this regard, this study is developed as a means of improving the diagnosis of Alzheimer’s disease through the application of transfer
learning to counter the challenges posed by scanty labeled data and excessive computational power required to train appropriate models in the medical imaging field. This study focuses on the
early classification of brain tumors using magnetic resonance imaging (MRI) and artificial intelligence (AI) through deep learning (DL). The researchers developed an efficient method based
on transfer learning, using pre-trained models such as Xception, NasNet Large, DenseNet121, and InceptionResNetV2 to extract deep features from MRI scans. The study utilized two benchmark
datasets, with preprocessing and data augmentation applied to enhance training. The models were trained using three optimizers (ADAM, SGD, RMSprop), and the Xception model with ADAM achieved
the best performance, showing 99.67% accuracy on the larger dataset and 91.94% accuracy on the smaller dataset, outperforming other approaches in the field. Our key contributions include: *
Use of transfer learning for MRI-based Alzheimer’s detection: we utilized three pre-trained deep CNNs—AlexNet, GoogLeNet, and MobileNetV2—applying transfer learning to leverage their
feature extraction capabilities while minimizing the need for large amounts of labeled training data. * Comprehensive performance evaluation: we conducted a detailed performance analysis,
assessing the effects of different solvers (Adam, Stochastic Gradient Descent, RMSprop) on model fine-tuning and classification accuracy. * Comparison of pre-trained architectures: we
compared these models in terms of accuracy, precision, and recall to determine the best-performing architecture for Alzheimer’s diagnosis using MRI datasets. Our analysis offers valuable
insights into the strengths and weaknesses of each model in the context of medical image classification. The rest of the paper is structured as follows: Sect “Materials and methods” details
the dataset we employed and the pre-processing techniques we applied. Sect “Results”offers a brief overview of previous studies. Sect “Limitations” discusses the proposed model and research
methods. Sect “Advantages” presents the experimental results and discussions, followed by the conclusion. RELATED WORKS In the last decade, the medical community has notably advanced imaging
techniques such as Positron Emission Tomography (PET), Diffusion Tensor Imaging (DTI), and particularly MRI, which has become a principal method for predicting AD25. For the classification
and diagnosis of AD, various deep learning techniques, such as support vector machines (SVM) using weighted MRI images and random forest classifiers, have been used26. El-Dahshan et al.27
suggested a hybrid approach involving feature extraction, classification, and dimensionality reduction, three-step classification techniques. This method is advantageous due to its speed,
simplicity, affordability, and non-invasive nature. However, a significant limitation of this hybrid technique is the requirement for new training whenever the dataset is increased. Ahmed et
al.28 introduced a CNN-based and patch-based classifier for classifying AD, reducing computing costs and significantly advancing the field. This approach involves multiple operations on the
dataset, enabling direct disease detection from the input data. In another study, Hong et al.29 developed a specialized recurrent neural network (RNN) based on long short-term memory (LSTM)
for the identification and classification of AD. This model uses current and historical patient data, making it crucial for timely disease identification. Furthermore, it primarily focuses
on predicting disease progression using time series data rather than just classification, making it more effective in anticipating future symptoms and enabling earlier interventions for
Alzheimer’s patients. This method uses the patient’s previous data that are connected to the patient’s present and historical data; this technique is significant for the timely
identification of the disease. The proposed technique primarily uses time series data and describes disease prediction instead of classification. Gupta et al.30 developed a model for
classifying AD using the ADNI dataset. Based on a sparse auto-encoder, the model achieves a maximum accuracy of 95%. Suk et al.31 introduced a new approach for classifying stages of AD and
Mild Cognitive Impairment (MCI) converters using an auto-encoder network, which achieved an accuracy of 95.9% across these MCI stages. Islam et al.32 proposed a CNN-based model for early
disease identification built on the Inception-V4 network. The input and output layers were processed using the inception-A, B, and C components and the reduction-A and B components, with
input and output passing through numerous filter concatenation procedures. The model achieved a total accuracy of 73.75% during the training and testing phases using the open-access imaging
studies (OASIS) database. A feature extraction framework was presented by Guerrero et al.33 that was established using substantial inter-subject variability. The proposed model achieved an
overall accuracy of 71%. However, it has some limitations due to its implementation as a binary classification model. Gorji et al.34 developed a convolutional neural network model with a
94.54% accuracy rate in classifying early and late stages of mild cognitive impairment. In Hosseini-Asl35, deep learning techniques have recently solved the inability of many computer-aided
diagnosis structures to extrapolate the distinguishing characteristics from the raw picture data automatically. There are four essential phases: features are extracted, segments are created,
skulls are stripped, and normalization is required in end-to-end learning to create an accurate prediction of diseases. Additionally, He et al.36 proposed the Deep Residual Network to
counteract the erosion of training accuracy, a common challenge exacerbated by the scarcity of annotated datasets. This limitation makes applying CNN in medical imaging particularly
challenging, as they require substantial data for practical training. Hazarika et al.37 examined several prominent deep-learning models and their effectiveness in classifying AD. Of these,
the DenseNet-121 model delivered the best performance. In another study, Shanmugam et al.38 tested three pre-trained CNN models (including GoogLeNet, AlexNet, and ResNet-18) for the early
detection of AD and related cognitive issues. Among them, ResNet-18 achieved the highest accuracy, recording a rate of 97.51%. Meanwhile, Battineni et al.39 utilized various deep neural
networks to classify MRI brain images, employing standard and fine-tuned features. This approach led to the highest accuracies of 95.7% with the CNN model and 95.5% with the DenseNet model.
Detecting Alzheimer’s disease (AD) through deep learning techniques has shown promise; however, several challenges persist that hinder the effectiveness and generalizability of these models
according to Table 1. Below is an analysis of how our research addresses the gaps in Alzheimer’s detection using MRI images through deep learning: Computational Resources and Efficiency:
Training deep architectures imposes significant computational resources and time overheads. A CNN-based patch classifier designed by Ahmed et al.,28 addresses these limitations with respect
to computational load, but the ensuing operations related to dataset’s creation are still elaborate and costly in resources, warranting the quest for more effective solutions. Our approach
addresses the challenge of computational burden by adopting transfer learning, which precludes the training of any models from scratch especially incurring low computational costs. Through
this strategy, it also becomes possible to make use of optimized/ trained CNN models which is an improvement from the previous work which were resource-intensive and involved complicated
processes. Additionally, we conducted a comparative analysis of three solvers (ADAM, SGD, and RMSprop), providing a comprehensive evaluation of their efficiency in terms of training
convergence and performance. This not only optimized model performance but also highlighted the most resource-efficient approaches, making our model more practical for real-world
applications where computational resources may be limited. Early detection and disease progression: while some approaches are devised that focus solely on identifying and classifying stages
of the disease after ample brain degeneration has taken place, early-stage detection is quite problematic. For the purpose of disease progression prediction, Hong and colleagues29 employed a
long-short-term-memory (LSTM)-based recurrent neural network, stressing this method’s scope was largely dependent on historical patient data made available, which can be a limitation for
early diagnosis. Our research effectively addresses the issue of the early detection of different stages of AD, including Very Mild-Demented and Mild-Demented ones. In other words, the model
can identify the early phase of dementia, which is essential to start intervention and treatment as soon as possible. This early detection is important as the progression of the disease can
be slowed down and the patient’s condition is likely to improve. By focusing on these early stages, our approach fills a key research gap, as identified in the work of Hong et al.,29 and
demonstrates the model’s capacity to support early-stage dementia identification, where interventions are most effective. Model Complexity and Interpretability: Accurate predictions have
been recorded in performing specific tasks using advanced models such as deep residual networks and autoencoders. However, such advanced models may present problems with regards to
interpretation and reproducibility. For example, Hosseini-Asl et al.35 proposed an end‐to‐end deep learning system for AD diagnosis but stressed that many preprocessing steps were necessary
adding to the complexity and potential sources of errors. In this research, we used CNN architectures like AlexNet, MobileNetV2, and GoogleNet that strike a good balance between performance
and interpretability as opposed to using complex models like autoencoders or deep residual networks. There is a degree of explainability which is intrinsic to CNNs as their convolutional
layers are looking at local features like edges or textures of MRI images and make it possible to see which areas of the brain contribute to the model. There is a high importance of this
form of interpretability in medical cases, as doctors need to know how the AI is making decisions. Thus, by applying pre-trained networks using transfer learning, we reduced computational
complexity while ensuring the model is simple and easy to understand. Limited dataset size and quality: deep learning architectures like convolutional neural networks (CNNs) rely on large,
annotated datasets to achieve their intended purpose effectively. Acquisition of such datasets is difficult in medical imaging due to the lack of labeled data as well as differences in
imaging protocols from one institution to the other. For example, He et al.36 pointed out that the availability of annotated data is a major bottleneck in training deep networks for AD
detection, especially so given that CNNs more often require large datasets. In our study, we efficiently incorporate transfer learning by incorporating pre-trained models (AlexNet,
MobileNetV2, and GoogleNet), which relieves the burden of large scale, well-annotated datasets. Datasets that are smaller in size can be used to fine-tune pre-trained models to address the
challenge of dataset scarcity. Moreover, the application of transfer learning in medical image analysis has gained much acceptance to address the challenge of limited dataset size.
Generalization across datasets: a model that is trained on specific data may not work as effectively on other data sets due to its unique distributions, imaging methods, or even different
patients. For instance, Battineni et al.39 achieved accurate results employing CNN and DenseNet architecture for Advanced Dementia diagnosis but indicated that some level of fine-tuning is
required to achieve such results efficiently, may limit generalizability across different MRI datasets. This work displays great results for the classification of AD with MRI images both on
the Kaggle dataset as well as OASIS. Nonetheless, we consider assessment of the model’s generalizability as very important, and we agree with the opinion of other studies, such as the one
conducted by Battineni et al.,39. Therefore, further research intends to incorporate external validation on different datasets to evaluate the performance of the model in various clinical
and imaging conditions. MATERIALS AND METHODS EMPLOYED DATASETS Using two MRI datasets, we tested our proposed model for identifying and classifying AD. The first dataset, available on
Kaggle40 and referred to here as the Kaggle dataset, is a multi-class, multi-label database containing 5000 images divided into four classes, each representing different stages of dementia
severity (Table 2). Given that the original images in the Kaggle dataset were highly imbalanced, which could lead to overfitting issues during deep learning training, we increased the number
of images. Different techniques were used to augment the data and improve model performance. Vertical and horizontal flipping, along with rotation, were applied to enhance the diversity of
the dataset. Initially, images were rotated by 45° to introduce variation. If class imbalance remained, additional rotations of 90 and 180° were performed. These combined techniques ensured
a balanced number of images across all classes, helping the model generalize more effectively. For the augmentation process, the same techniques were applied uniformly across all classes to
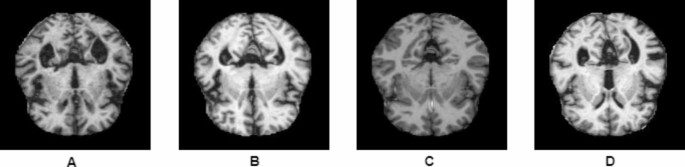
maintain consistency. This approach successfully expanded the Kaggle dataset to 10,254 MRI images (Table 2; Fig. 1). Figure 1 shows sample images for each class from the Alzheimer’s disease
dataset. The images were resized to meet the input specifications of the pre-trained models. We also performed the external evaluation of our proposed model using a second dataset of 382 MRI
images sourced from the OASIS database curated by Daniel S. Marcus at the Neuroimaging Informatics Analysis Center (NIAC) at Washington University School of Medicine41 (Table 3). This
dataset, referred to here as the OASIS dataset, consists of 3 to 4 individual T1-weighted MRI scans contributed by each subject, all obtained during a single imaging session. For
classification purposes, we divided the images into four classes according to the (CDR) scores: CDR-0 (non-demented), CDR-0.5 (very mild-demented), CDR-1 (mild-demented), and CDR-2
(moderate-demented). This dataset comprises MRI images of AD patients aged 23 to 86 years. To ensure balanced data and enhance the model’s learning efficacy, we also applied an augmentation
strategy to this dataset. While the original OASIS41 dataset images are 256 × 256 pixels, our proposed CNN model requires images to be resized to 224 × 224 pixels. Therefore, we
appropriately scaled the OASIS dataset images to fit the required dimensions for our model. Finally, for both datasets, we partitioned the data into two subsets: the training set, which
comprises 80% of the images, and the test set, which contains the remaining 20% to provide sufficient training data, ensure better generalization, and balance overfitting and underfitting.
Although the Kaggle dataset is popular, it had problems with class imbalance and a limited number of images, which were fixed during model training. We evaluated the trained model’s
generalizability using the OASIS dataset for external validation, and it performed admirably. Our study is currently restricted to these two datasets, but we intend to test the models on
more datasets in the future to assess their generalizability and performance in further detail. GRAD-CAM VISUALIZATIONS Figure 2 presents the Grad-CAM Visualizations of Correctly MRIs.
Original MR images of four different types alongside their respective Grad-CAM heatmaps. Each pair shows the original image on the left, followed by the Grad-CAM visualization on the right.
The true and predicted classes are displayed for each image. Grad-CAM highlights the regions of the image that the model focused on when making its classification decision. Warmer colors
(red/yellow) indicate regions with a higher contribution to the model’s decision, while cooler colors (blue/green) signify less impact. These visualizations provide insight into the model’s
attention to key features that align with expert-based classification criteria. Figure 3 shows the Grad-CAM Visualizations of Misclassified MRIs. Misclassified MRIs of four Alzheimer’s
classes and their corresponding Grad-CAM heatmaps. For each pair, the left image shows the original MR image, while the right image displays the Grad-CAM heatmap. The true class and
predicted class for each image are labeled, providing insights into the model’s misclassification patterns. Grad-CAM highlights the regions of the image that the model focused on during its
prediction process, with warmer colors (red/yellow) indicating areas that contributed most to the model’s decision, and cooler colors (blue/green) showing less influential regions. These
visualizations reveal potential challenges in differentiating between MR images of Alzheimer’s types. EXPERIMENTAL SETUP This study’s deep neural networks were developed using Matlab version
9.8, 2020a. All experiments were conducted on a machine with an Intel Core (TM) i7-6500 CPU @ 2.50 GHz and 16 GB RAM. In this research, we applied CNN pre-trained neural networks through
transfer learning to classify AD. This research utilizes pre-trained models such as AlexNet, GoogleNet, and MobileNetV2 for classification purposes. We carefully examined how changing
different parameters could help us achieve the best accuracy for our model. We experimented with various epochs and different initial learning rates during training to understand their
impact on performance. After that, we investigated which solver would work best with the parameters we tested. To do this, we tried out several solvers with three different architectures.
Our goal was to fully fine-tune the model, ensuring it performs optimally based on our findings. Table 4 shows the architectures used in the study, the solvers applied, initial learning
rates, and the number of training epochs. AlexNet, GoogleNet, and MobileNetV2 all used SGDM, ADAM, and RMSPROP as solvers, with a default batch size of 128. The initial learning rate is set
to 0.001, and each model was trained for 25 epochs. Since these pre-trained models have already undergone training on extensive databases of images, they require significantly less time and
computing resources for further training than developing new models45. These networks are comprised of thousands of neurons and have millions of parameters. AlexNet, with 62.3 million
parameters, pioneered deep learning with its three fully connected and five convolutional layers, while GoogleNet’s 22 layers, expanded to 27 with pooling layers, revolutionized neural
network architectures. MobileNetV2, with fifty-three deep layers, excels in classifying 100 object categories, leveraging millions of images for training. Although the number of layers in
each model differs, some common layers are found in almost all CNN models, including input, convolution, pooling, fully connected, and output layers42. The solver coordinates the network’s
forward inference and backward gradients to create parameter updates that aim to reduce the loss. This process is known as model optimization. The solver is in charge of managing the
optimization and producing parameter updates. In our study, we used three different solvers to optimize training architectures, as follows43: STOCHASTIC GRADIENT DESCENT WITH MOMENTUM (SGDM)
Stochastic gradient descent with momentum (SGDM) is an optimization algorithm that enhances the basic gradient descent approach by introducing a momentum factor. SGDM updates the network’s
weights and biases to reduce the loss function by incrementally moving toward the negative gradient of the loss function with each iteration. This momentum factor helps reduce oscillation
during parameter updates, leading to smoother convergence towards the optimal solution43. $$\delta \ell +1\,=\,\delta \ell - a\nabla E(\delta \ell ){\text{ }}+g(\delta \ell - \delta \ell -
1)$$ (1) > Where ℓ represents the number of iterations, α > 0 specifies > the learning rate, δ represents the parameter vector, E(δ) denotes > the loss function, and γ indicates
the contribution of the previous > gradient step to the current iteration. ROOT MEAN SQUARE PROPAGATION (RMSPROP) RMSPROP is one method that stores a moving average using the squares of
the parameter gradients by element43. $$v\ell ={\rm B}2v\ell - 1+(1 - {\rm B}2){\text{ }}[\nabla E(\delta \ell )]2$$ (2) vℓ: Exponentially Weighted Moving Average (EWMA) of the square of
gradients at layer ℓ. B2: Hyperparameter scaling the previous EWMA term. (1−B2): Complement of B2, controlling direct contribution of current gradient’s square. [∇E(δℓ)]^2: Square of the
gradient of the loss function. Multiplying B2 with vℓ scales the previous EWMA term. Multiplying (1−B2) by [∇E(δℓ)]^2 scales the current gradient’s square. Combined, they update parameters
during optimization, often used in algorithms like Adam for stochastic optimization. Where B2 reflects the moving average’s decay rate. The squared gradient’s average length is equal to 1/
(1−B2), with the value of 10, 100, and 1000 parameter updates, respectively43. $$\delta \ell +1\,=\,\delta \ell - a\nabla E(\delta \ell ){\text{ }}/\surd v\ell + \in$$ (3) Here, each element
is divided separately When using RMSProp, the learning rates for parameters with small gradients are effectively increased, while the learning rates for parameters with high gradients are
effectively decreased. To prevent division by zero, a small constant ɛ is included. ADAPTIVE MOMENT ESTIMATION (ADAM) ADAM employs a parameter similar to RMSPROP with the addition of a
momentum term. It maintains track of the parameter gradients and their squared values using a moving average based on elements43. $$m\ell = {\rm B}1m\ell - 1 + (1 - {\rm B}1)\nabla E{\text{
}}(\delta \ell )$$ (4) $$v\ell =B2v\ell - 1+(1 - B2){\text{ }}[\nabla E(\delta \ell )]2$$ (5) Where the decay rates for the training choices squared gradient decay factor and gradient decay
factor are B1 and B2, respectively. To update the network settings, Adam employs moving averages as43: $$\delta \ell +1\,=\,\delta \ell - aml/\surd vl+\smallint$$ (6) Where ɛ with the help
of the epsilon training option. If gradients across numerous iterations are the same, using a moving average of the gradient enables parameter changes to build momentum in one direction.
When noise dominates the gradients, the gradient’s moving average becomes smaller, resulting in smaller parameter updates. The proposed model is shown in Fig. 4. The diagram represents a
comprehensive workflow for proposed model, divided into three key layers: data layer, training layer, and testing layer. Detailed explanation of each layer is given as: DATA LAYER Data
Source: Data is collected from hospitals, consisting of Alzheimer’s patient scans. Data Augmentation & Image Processing: To improve the robustness of the model, data augmentation
techniques were implemented such as flipping, rotation, scaling, etc. This step helps in creating more diverse training data and prevents overfitting. Image processing techniques also
include that the images are converted into a suitable format for the model. Data Division: After preprocessing, the dataset is divided into training, and testing sets to ensure proper
evaluation of the model’s performance. TRAINING LAYER Model selection: Three different pre-trained convolutional neural networks (CNNs) are utilized: AlexNet, GoogleNet, and MobileNetV2.
These models are known for their capability to extract significant image features. Optimizer choice: Three different optimization techniques like SGDM, ADAM, and RMSPROP are used for each
architecture. These techniques adjust the model’s weights during training to make the model perform better. Learning criteria: The process involves a decision block to determine whether the
model has met the predefined performance criteria. If the model does not meet the expected accuracy or other criteria, it loops back for weight retraining, optimizing until satisfactory
results are achieved. TESTING LAYER The testing phase involves using unseen images to evaluate the model’s generalization capability. Classification: The trained model classifies the input
images into one of the four categories: mild demented, moderate demented, very mild demented, and non-demented. This classification assists in detecting the level of Alzheimer’s disease in
the patient based on the provided medical images. The evaluation matrices listed below were employed to assess the performance of the implemented models7: $$Accuracy~=\frac{{\varepsilon +
\propto }}{{\varepsilon +\gamma + \propto +\delta }}$$ (7) $$Specificity~=\frac{ \propto }{{ \propto +\gamma }}$$ (8) $$\operatorname{Re} call~or~Sensitivity=\frac{\varepsilon }{{\varepsilon
+\delta }}$$ (9) $$\Pr ecision~=\frac{\varepsilon }{{\varepsilon +\gamma }}$$ (10) $$F1~score~=\frac{{2*~\Pr ecision~*~\operatorname{Re} call~}}{{~\Pr ecision~+~\operatorname{Re} call~}}$$
(11) $$Misclassification~rate=100~ - ~(\frac{{\varepsilon + \propto }}{{\varepsilon +\gamma + \propto +\delta }})$$ (12) Where ε represents the predicted true positive value, ∝ represents
the predicted true negative value, γ represents the predicted false positive value, and δ represents the predicted false negative value. RESULTS For classification purposes, we divided the
images into four classes according to the CDR scores: CDR-0 (Non-Demented), CDR-0.5 (Very Mild-Demented), CDR-1 (Mild-Demented), and CDR-2 (Moderate-Demented). We implemented three distinct
CNN models (AlexNet, GoogleNet, and MobileNetV2), testing each model with three different solvers (SGDM, ADAM, RMSPROP) to identify which model performs best with each solver. In this
section, we provided detailed results for each model alongside the solver it performed best with, ensuring that the results section is brief. Table 5 illustrates the number of epochs,
initial learning rate, number of iterations, implemented solvers, and accuracy performance of each model. We calculated the true positive, true negative, false positive, and false negative
values for each class using the models that yielded the best results with specific solvers. Based on these values, we performed additional calculations to assess the implemented
architectures’ class-wise precision, misclassification rate, accuracy, recall, specificity, and F1 score. After selecting the optimal hyper-parameters, we evaluated the performance of these
models using all the data from the training and testing subsets CLASS-WISE DETAILED RESULTS OF IMPLEMENTED ARCHITECTURES TRAINED WITH OPTIMAL SOLVERS PERFORMANCE EVALUATION OF ALEXNET MODEL
TRAINED WITH ADAM SOLVER To evaluate the effectiveness of different CNN architectures, we tested the AlexNet model using the ADAM solver on a dataset of MRI images. We assessed the
performance of the AlexNet model trained with the ADAM solver on four classes of MRI images sourced from the Kaggle dataset. Our findings revealed that the model attained an accuracy of
99.4%. We evaluated the class-wise precision and recall of the model (Table 6; Figs. 5 and 6). Our findings revealed that the model achieved 99.4% precision and 100% recall for the
Mild-Demented class, 100% precision and 100% recall for the Moderate-Demented class, 99.6% precision and 98.0% recall for the Non-Demented class, and 98.6% precision and 99.6% recall for the
Very Mild-Demented class. Next, we assessed the number of correctly classified images in each class. Out of the 515 images in the Mild-Demented class, 512 were correctly classified, with
three misclassified. All 514 images in the Moderate-Demented class were correctly classified. Of the 504 images in the Non-Demented class, 502 were correctly classified, with two
misclassified. Lastly, out of the 518 images in the Very Mild-Demented class, 511 were correctly classified, with seven misclassified. Analyzing the performance of the AlexNet model, which
was trained with the ADAM solver, across various stages of Alzheimer’s Disease, namely: mild-demented, moderate-demented, non-demented and very mild-demented disorders has also been done in
Table 6. Performance was quantified in terms of Accuracy, Precision, F1-Score, Recall, Specificity and Misclassification Rate, for the respective cases. While the model performed quite well,
we noticed very few instances of misclassification especially in the Non Demented and Very Mild Demented levels. The model performed exceptionally well for the Mild-Demented and
Moderate-Demented classes, with the Moderate-Demented classification showing 0% misclassification, indicating the accuracy of the model in differentiating moderate dementia from other
levels. In contrast, there were very low (0.00585 and 0.00439, respectively) misclassification rates for the Non-Demented and Very Mild-Demented classes, which indicate that it may be
somewhat difficult to the subject who can carry the two stages. More specifically in the Non-Demented class, there were 10 False Negatives (FN) and 2 False Positives (FP), which means that
the Recall (98.05%) and F1-Score (98.82%) were slightly low than in other classes. This could mean that some Non-Demented individuals have dementia stage features overlapped with them which
makes it difficult for the model to distinguish between very early dementia, and completely healthy individuals, as is the case here. Subtle structural changes in the brain that do not
manifest as clinical symptoms may contribute to these misclassifications. There were two False Negatives (FN), and seven False Positives (FP) in the Very Mild-Demented class, which provided
a Precision of 98.65 and a Recall of 99.61. The higher rate of False Positives in this class indicates that the model at times, misclassified Non-Demented patients as those with Very
Mild-Dementia. This may be attributed to the thin line that exists between normal aging and a very mild form of cognitive impairment that exhibits no structural changes in the brain at this
stage — changes that can easily be confused with variations within normal aging patterns. Possible Reasons for misclassifications could be the feature overlap– Non-Demented and Very mild
Demented individuals tend to have indistinct MRI characteristics in most cases as in the case of atrophy or thinning of the hippocampus. Entering into some of these changes in the brain at
this stage is a feature that may not be easy to appreciate without finer techniques of feature extraction. CNNs are great for facilitating the process of feature extraction. When it comes to
MRI scans, there are usually some finer points that unfortunately this model fails to capture most especially for cases that border on the extremes. Transferring learning might be effective
but there would be need for more tweaks or more complex architectures to enhance the results. All calculated values described in Table 6 are shown graphically in Fig. 7. PERFORMANCE
EVALUATION OF GOOGLENET MODEL TRAINED WITH ADAM We assessed the performance of the GoogleNet model trained with the ADAM solver on four classes of MRI images sourced from the Kaggle dataset.
Our findings revealed that the model attained an accuracy of 98.0%. We evaluated the class-wise precision and recall of the model (Table 7; Figs. 8 and 9). Figure 9 demonstrates that we
attained a precision of 99.8% and a recall of 100% for the Mild-Demented class, 100% precision and recall for the Moderate Demented class, 92.6% precision and 100% recall for the
Non-Demented class, and 100% precision and 91.8% recall for the Very Mild Demented class. Of the 513 images in the Mild-Demented class, 512 were correctly classified, with one misclassified.
All 514 images in the Moderate-Demented class were correctly classified. Of the 533 images in the Non-Demented class, 512 were correctly classified, with 41 misclassified. Lastly, all 471
images in the Very Mild-Demented class were correctly classified. Next, we assessed the class-specific GoogleNet trained with ADAM architecture’s performance according to Accuracy,
Precision, F-1 Score, Recall, Specificity and Misclassification rate (Table 7; Fig. 10). For the Mild-Demented class, the following results were attained: Accuracy of 99.95%, F-1 score of
99.90%, Precision of 99.81%, Specificity of 99.94%, Recall of 100%, and a Misclassification rate of 0.000487567. For the Moderate-Demented class, the model achieved an Accuracy, F-1 score,
Precision, Specificity, and Recall of 100%, with a 0 Misclassification Rate. For the Non-Demented class, the following results were obtained: Accuracy of 98.0%, F-1 score of 96.15%,
Precision of 92.59%, Specificity of 97.34%, Recall of 100%, and a Misclassification rate of 0.019990249. For the Very Mild-Demented class, the results were as follows: Accuracy of 97.95%,
F-1 score of 95.73%, Precision of 100%, Specificity of 100%, Recall of 91.81%, and a Misclassification Rate of 0.020477816. The model demonstrated exceptional performance across different
dementia classes, achieving high accuracy, precision, recall, and specificity. Table 7 illustrates the performance results obtained with the GoogleNet model trained using the ADAM solver for
a four-stage AD classification namely Mildly-Demented, Moderately-Demented, Non-Demented and Very Mildly-Demented too. Although the model performed well in accuracy, precision, recall and
specificity on most of the classes, there are some crucial points of the class misclassification rates, especially in Non-Demented and Very Mild-Demented classes, that should be addressed.
As for the Mild-Demented and Moderate-Demented classes, the model performed almost perfectly, achieving a recall and precision of 1, which implies that the model can easily differentiate the
above stages from the rest. The misclassification rates are close to zero, The Non-Demented category, on the other hand, had 41 False Positives, which affected the precision to 92.59%,
which means that some Non-Demented persons were labeled as demented. Such a misdiagnosis can have implications in healthcare as it can lead to needless interventions or treatments for
patients who are wrongly diagnosed with dementia. Even though there is no dementia case that is in this class that was left out, as indicated by the False Negative rate of 0%, the alarming
False Positive rate (41 images) compromises the precision increasing chances of overdiagnosis. In a real-life situation, this introduces frustration to both the patients and their relatives
as well as unnecessary follow-ups or treatments. The class of very mild-demented had 42 cases (False Negatives) which were wrongly identified bringing the recall at 91.81%. Recall here
suggests that the model failed to determine 42 cases of very mild dementia. When it comes to the precision, it stands at 100%, that is, No Non-Demented patient was misclassified as very
mild-demented. However, the low recall shows that the model was able to identify almost 8% of patients who were very mild-demented. For example, this type of underdiagnosis can postpone
treatment, therefore the patients may not benefit from early treatment which is fundamental to curbing the advancement of AD. The Very Mild-Demented class presents a paradox; this is an
instance where the model manages to detect cases but with some limitations. High precision means that when the model classifies an instance as a case of very mild dementia, it is accurate
100% of the time. Nonetheless, the recall is relatively low (91.81%) and this indicates that there are many actual cases that go undetected which is quite worrisome in a clinical setting
where one aims to detect the onset of the illness as early as possible i.e. AD. The False Negatives within this level illustrate losses in regard to identifying the disease at an appropriate
stage, hence, allowing room for treatments which may not be helpful to the patient. PERFORMANCE EVALUATION OF MOBILENETV2 MODEL TRAINED WITH SGDM SOLVER > We assessed the performance of
the MobileNetV2 model trained with > the SGDM solver on four classes of MRI images sourced from the > Kaggle dataset. Our findings revealed that the model attained an > accuracy of
96.5%. We evaluated the class-wise precision and recall of the model (Table 8; Fig. 11, and Fig. 12). Figure 11 illustrates that we achieved 93.43% precision and 100% recall for the
mild-demented class, 99.8% precision and 100% recall for the moderate-demented class, 100% precision and 86.5% recall for the non-demented class, and 93.8% precision and 99.6% recall for the
very mild-demented class. Of the 548 images in the mild-demented class, 512 were correctly classified, with 36 misclassified. Of the 515 images in the moderate-demented class, 514 were
correctly classified, with one misclassified. All 443 images in the very non-demented class were correctly classified. Lastly, out of the 545 images in the very mild-demented class, 511 were
correctly classified, with 34 misclassified. Next, we assessed the class-specific MobileNetV2 trained with SGDM architecture’s performance according to Accuracy, Precision, F1-Score,
Recall, Specificity, and Misclassification rate (Table 8; Fig. 13). For the Mild-Demented class, we achieved 98.24% Accuracy, 93.43% Precision, 96.60% F1 score, 100% Recall, 97.66%
Specificity, and a Misclassification rate of 0.017552413. For the Moderate-Demented class, we achieved 99.95% Accuracy, 99.81% Precision, 99.90% F1 score, 100% Recall, 99.93% Specificity,
and a Misclassification rate of 0.000487567. For the Non-Demented class, we attained an Accuracy of 96.64%, a Specificity of 100%, an F1 score of 92.77%, a Recall of 86.52%, a Precision of
100%, and a Misclassification rate of 0.033642126. Moreover, for the Very Mild-Demented class, we achieved 98.24% Accuracy, 93.76% Precision, 96.60% F1 score, 99.61% Recall, 97.79%
Specificity, and a Misclassification rate of 0.017552413. Table 8 shows how the MobileNetV2 model was evaluated in four stages of dementia: mild-demented, moderate-demented, non-demented,
and very mild-demented. The system performs well at 99.66% and 97.31% accuracy rates in the moderate-demented and non-demented classes respectively. However, several errors of classification
are present in the mild-demented and the very mild-demented classes which need to be evaluated, understood, and resolved. When it comes to the mild-demented class, it shows a 1.76%
misclassification rate with 36 false positives (FP) leading to a Precision of 93.43% and an F1-Score of 96.60%. Likewise, when it comes to the Very Mild-Demented class, it also demonstrates
a 1.76% misclassification rate with 34 False Positives (FP) and 2 False Negatives (FN) giving rise to 93.76% precision and an F1-Score of 96.60%. The aforementioned misclassifications
indicate that the model experiences difficulty differentiating between the early stages of dementia, such as Mild and Very Mild Demented, and other stages. This may be due to the absence of
distinct brain changes since these changes start in milder forms of dementia which may also be present in the elderly or in the more advanced forms of dementia. Characteristics observed via
MRI may include atrophy of the hippocampus or thinning of cortical levels which may not be visibly pronounced; causing the model to confuse Mild-Demented and Very Mild-Demented patients with
those who do not have dementia but are in a more advanced stage. In the classification of dementia, the problem of class overlaps in the early-stage diagnosis is a serious one, as it is
often the case that changes in the brain are subtle or non-specific. Here, the term False Positives point to the tendency of the model to incorrectly label Non-Demented subjects as
Mild-Demented and Very Mild-Demented individuals. In practice, this might lead to an increase in the errors of diagnosis among the population, including the treatment of patients with normal
cognition. The model performs exceptionally well in the Moderate-Demented class recording an Accuracy of 99.95%, Precision of 99.81%, and a mere 0.05% misclassification rate. This implies
that the model is quite effective in classifying more advanced forms of dementia as there are gross changes in the brain that can be easily identified. In contrast, the Non-Demented class
shows a rate of 3.36% of misclassification with 69 False Negatives (FN) recorded which resulted in Recall of 86.52% and F1-Score of 92.77% respectively. The False Negatives in this class may
suggest that some Non-Demented individuals are being. Using the precision and recall metrics it is possible to draw conclusions about the clinical relevance of the model. For example, high
precision means that when the model classifies a case as positive Mild-Demented or Very Mild-Demented, this is most likely to be correct, which is important in avoiding becoming overzealous
in pursuit of a diagnosis where one does not exist that may lead to harmful treatments. The non-demented and mildly demented categories being associated with lower recall means that, in
fact, the model may be misclassifying some early-stage patients, which would lead to some cases being undiagnosed. In real-world clinical practice, it becomes imperative for a physician to
be able to detect dementia among patients since it is known that early intervention would mitigate the course of the disease. The model exhibits low recall in the Non-Demented and Very
Mild-Demented classes, which implies that some cases of early-stage dementia in practice may go undetected, leading to a postponement in treatment. This is particularly true here where
improving recall in these classes is very important to maximize the detection of cognitive impairment at its most early stages for timely intervention. CLASS-WISE PERFORMANCE SUMMARY FOR ALL
ARCHITECTURES As shown in Table 9, the AlexNet, GoogleNet, and MobileNetV2 models were evaluated across four classes: mild demented, moderate demented, non demented, and very mild demented.
For Mild Demented, all models had good results, and all the models were examined to have perfect recall, and minimized misclassification were noticed but precision of MobileNetV2 was not
much better, which was 93.43%. In Moderate Demented, both AlexNet and GoogleNet performed perfectly across all metrics (100%), and MobileNetV2 also showed strong performance but with a
slight misclassification rate. In non Demented, overall results are encouraging but recall results of MobileNetV2 and Precision of GoogleNet are slightly low. In Very Mild Dementia, it was
found that GoogleNet had perfect precision but low recall (91.81%), while AlexNet had excellent precision (98.65%) and high recall (99.61%). EXTERNAL EVALUATION PERFORMANCE OF THE PROPOSED
MODEL ON THE OASIS DATASET Our experiments revealed that the AlexNet model trained with the ADAM solver achieved the highest performance. To validate this finding, we conducted an external
evaluation to assess the performance of this model using four classes of MRI images from the OASIS dataset. We found that the model achieved 98.2% classification accuracy on the OASIS
dataset using the AlexNet architecture and ADAM solver. We evaluated the model’s class-wise precision and recall (Figs. 14 and 15). Our findings revealed that the model achieved 100%
precision and recall for the mild-demented class and 100% precision and recall for the moderate-demented class. Additionally, it achieved 100% precision and 94.0% recall for the Non-Demented
class and 92.4% precision and 100% recall for the very mild-demented class. We evaluated the class-specific proposed model’s performance according to accuracy, precision, F1- score, recall,
specificity, and misclassification rate (Table 10; Fig. 11). For the mild-demented class, we achieved 100.00% Accuracy, 100.00% Precision, 100.00% F1-Score, 100.00% Recall, 100.00%
Specificity, and a Misclassification Rate of 0. For the Moderate-Demented class, we achieved 100.00% Accuracy, 100.00% Precision, 100.00% F1-Score, 100.00% Recall, 100.00% Specificity, and a
Misclassification Rate of 0. For the Non-Demented class, we achieved 98.18% Accuracy, 100.00% Precision, 96.91% F1 Score, 94.01% Recall, 100.00% Specificity, and a Misclassification Rate of
0.018214936. For the Very Mild-Demented class, we achieved 98.24% Accuracy, 92.42% Precision, 96.06% F1-Score, 100.00% Recall, 97.76% Specificity, and a Misclassification Rate of
0.017574692. The class-specific proposed model demonstrated remarkable performance across various dementia classes, achieving high accuracy, precision, recall, and specificity. In Fig. 16,
each bar reflects the corresponding metric for each class, with 100.00% values for most metrics in mild demented and moderate demented classes, and slightly lower values for non-demented and
very mild demented. Specifically, non-demented has a Recall of 94.01% and F1 Score of 96.91%, while Very Mild Demented shows a Precision of 92.42% and F1 Score of 96.06%. PERFORMANCE
COMPARISON OF PROPOSED MODEL ON MULTIPLE DATASETS In this research paper, we proposed a novel approach for early detection of Alzheimer’s disease using MRI scans and transfer learning
techniques with pre-trained CNNs. Our study focused on three distinct CNN architectures, AlexNet, GoogleNet, and MobileNetV2, trained using three different solvers: SGDM, ADAM, and RMSPROP.
We tested the models on two datasets: the Kaggle dataset and the OASIS dataset. The choice of solver significantly influenced the performance of the models, as the ADAM solver consistently
yielded the highest accuracy across all architectures. SGDM and RMSPROP also produced competitive results but generally lagged behind ADAM. The AlexNet model achieved the highest accuracy of
99.4% on the Kaggle dataset when trained with the ADAM solver as shown in Table 10, followed by the GoogleNet model with an accuracy of 98.2% and the MobileNetV2 model with an accuracy of
96.5%. The class-wise evaluation of the models revealed their ability to classify different stages of Alzheimer’s disease accurately. In our research, we identified several constraints,
including the dataset size, and the model’s generalizability to different datasets. To enhance our evaluation, we utilized an external dataset called OASIS, which allowed us to assess how
well our proposed model performs on unknown MRI images. The results from this new dataset were encouraging. We compared our findings with the previous studies, which utilized the same
datasets from Kaggle and OASIS as shown in Tables 11 and 12. Our results were indeed encouraging. The results align closely with the expected outcomes, showcasing the model’s effectiveness.
The performance can be significantly enhanced by meticulously fine-tuning the model, adjusting parameters such as the number of epochs and initial learning rate, and, most importantly,
identifying the optimal solver. In the context of Alzheimer’s disease detection, the ADAM optimizer stands out due to its ability to adaptively adjust the learning rate for each parameter.
This feature enhances the model’s ability to learn from complex data, such as MRI images, by efficiently handling sparse gradients that often occur in medical imaging tasks. The proposed
model is designed to assist doctors in automatic disease detection. These findings can enhance clinical Alzheimer’s disease detection by providing more accurate and reliable models for
identifying the disease in patients. Improved detection methods can lead to earlier diagnosis and better treatment strategies. LIMITATIONS While our proposed approach for Alzheimer’s disease
detection using transfer learning and pre-trained CNN models has shown promising results, several limitations must be acknowledged. Our models rely on pre-trained networks, which were
initially trained on large-scale datasets unrelated to medical images. This may result in a suboptimal feature extraction for MRI data compared to training CNNs from scratch on medical
datasets, although the latter would require much larger labeled datasets. Although the model performed well on the Kaggle and OASIS datasets, the generalizability of the model to other MRI
datasets remains untested. MRI data from different sources might vary due to differences in imaging protocols, patient demographics, and scanner settings, which could affect model
performance. Despite using transfer learning to reduce the computational cost, the fine-tuning process still requires significant computational power and time for training, especially when
testing different solvers and models. Although the OASIS dataset is a useful tool for detecting Alzheimer’s disease, it contains built-in restrictions that could affect how broadly
applicable, our findings are: * 1. 1. Imaging techniques: The standard techniques used to capture images in this database may differ from those used in other imaging datasets or in clinical
practice. When evaluated on fresh and varied datasets, models may perform differently due to variations in the SUBJECTS involved, such as scanners, resolution, and imaging sequences. * 2. 2.
Demographic variability: When using a resource like OASIS, researchers are limited to using data taken from a group of people who fall into particular age and geographic categories. This
could introduce bias into the model, making it potentially inaccurate. To address these limitations and improve the robustness of the model, we have taken the following steps: * 3. 1. Use of
multiple datasets: yo provide preliminary external validation, we tested the model’s performance across various imaging methods and demographics using two datasets (Kaggle and OASIS). * 4.
2. Transfer learning with pretrained models: to take use of a variety of features and optimization strategies, we used transfer learning with three pretrained models (AlexNet, GoogleNet,
MobileNetv2) and solvers (ADAM, SGDM, and RMSprop). ADVANTAGES Our research provides several key advantages that distinguish it from previous work in Alzheimer’s disease detection. Our use
of transfer learning with fine-tuned CNN architectures resulted in a high classification accuracy, with AlexNet achieving 99.4% on the Kaggle dataset, outperforming several state-of-the-art
models. By utilizing pre-trained models, we significantly reduced the need for extensive computational resources and time, compared to training models from scratch. This method is quicker to
put into action and is more geared towards researchers without any available resources. We compared the effectiveness of different solvers (ADAM, SGDM, RMSprop) with three different CNN
architectures and showed how solver selection helps in improving the performance of the model. This method could be modified to suit other medical image classification tasks apart from the
detection of Alzheimer’s disease, hence making it quite useful in medical diagnostics. BROADER IMPACT STATEMENT The proposed model has significant implications for real-world clinical
applications, particularly in under-resourced settings: * 1. Adoption in resource-limited environments: the suggested model’s computational simplicity, which is attained by combining
transfer learning with lightweight architectures like MobileNetv2, makes it ideal for deployment in environments with constrained computational resources. Utilizing pretrained models reduces
the requirement for intensive local training, which further reduces computational expenses. * 2. Scalability and accessibility: the model can be used in a variety of clinical settings,
including ones with varying imaging methods and demographic characteristics, as evidenced by its flexibility with regard to different datasets. This scalability guarantees that a variety of
healthcare facilities can use the system. * 3. Reducing barriers to adoption: the model’s open-source framework and low computational needs enable its adoption in resource-constrained
environments, allowing healthcare providers to implement it without major infrastructure upgrades. * 4. Future directions for impact: in order to facilitate clinical acceptance, future
research endeavors may concentrate on developing intuitive user interfaces and incorporating the model into current diagnostic procedures. Furthermore, working together with physicians in
contexts with limited resources may yield insightful input that helps improve the system for real-world use. These considerations highlight the potential of the model to make a meaningful
impact in real-world healthcare. FUTURE WORK The work presented here may be developed in several ways in order to make it more solid and applicable to other patient groups. Future studies
should proceed to the verification of the models proposed on other MRI datasets from other centers and countries in order to evaluate the efficacy and applicability of the model in various
imaging settings. In addition to MRI, imaging modalities such as PET or CT can be combined in a muli-modal deep learning framework which will help increase the sensitivity of diagnosing
Alzheimer’s disease by adding more information through other imaging techniques. Implementation of more modern deep learning architectures such as EfficientNet or Vision Transformers (ViTs)
are also possible to enhance the accuracy level and further cut down the training period since they are efficient and highly scale up for images classification. Future work could focus on
optimizing the model for real-time applications in clinical settings, where time efficiency and interpretability are critical for providing timely diagnoses. CONCLUSION This research
presented a new concept in classifying Alzheimer’s disease (AD) in four classes, namely mild-demented, moderate-demented, non-demented, and very mild-demented. Employing transfer learning
and adjusting three pre-trained convolutional neural networks AlexNet, MobileNetV2, and GoogleNet, the classification accuracy was very high with the pre-trained AlexNet model with ADAM
solver achieving the highest accuracy. This high accuracy suggests that our approach could facilitate earlier and more accurate diagnosis of Alzheimer’s disease, which is critical for timely
intervention. Earlier detection of AD allows for prompt treatment and management strategies, potentially improving patient outcomes by slowing disease progression. The outcomes of this
research have substantial clinical implications. For example, very high classification accuracy in the earlier stages of dementia, such as Very-Mild-Demented and Mild-Demented, implies that
such a model may be operationalized in the clinical settings to assist medical practitioners in making an accurate and timely diagnosis of Alzheimer’s disease prior to what is currently
possible, thereby improving the treatment interventions offered. Whereas AD is at its most challenging to diagnose at incipient stages due to various factors, the present approach provides a
consistent and dependable approach that minimizes the chances of misdiagnosis and helps in preventing and improving the treatment of various disorders, hence increasing the well-being of
patients. Our methodology, in addition to Alzheimer’s disease, seems to have a good purpose for additional medical conditions, mostly due to the characteristics of medical imaging. This is
because transfer learning can easily incorporate any pre-trained models, which makes it possible for use even in other degenerative conditions likely Parkinson’s disease or even various
other imaging techniques such as CT or PET scans. Further fine-tuning would be needed to adapt the models to the specifics of each condition and imaging modality, but the general methodology
remains applicable across various healthcare diagnostics. In relation to the current methodologies in the field, our model boasts remarkable benefits in terms of both precision and
processing time. Because we employed transfer learning technique rather than training models afresh, we cut down operational expenses and yet comparable accuracy was retained. This not only
guarantees less time consumption when making the models but also makes the incorporation of artificial intelligence in medicine less challenging to the physicians who have little computing
power. Given the superior performance coupled with ease of use, it is conceivable that our model will perform better than the conventional machine learning models, which depend on feature
engineering and large data sets. As we improve our approach, the next steps in the work will concentrate on cross-dataset generalization of the models, enhancing potential analysis of the
models through explainable AI, and implementing real-time clinical deployment to increase diagnostic capabilities and enhance patient care. DATA AVAILABILITY The dataset and simulation files
used during the current study are available from the corresponding author upon request. CODE AVAILABILITY Current code version: V1.0, Permanent link to code/repository used for this code
version: https://github.com/KainatSaher/Alzheimer/blob/main/task.m, Software code languages, tools and services used: Matlab, Compilation requirements, operating environments, and
dependencies: Core i7-9850H Precision 5540, 32GB RAM, Microsoft Windows, Support email for questions: [email protected] and [email protected]. REFERENCES * Mehmood, A. et al. A
transfer learning approach for early diagnosis of Alzheimer’s disease on MRI images. _Neuroscience_ 460, 43–52 (2021). Article CAS PubMed MATH Google Scholar * Association, A.
Alzheimer’s disease facts, and figures include a special report on Alzheimer’s detection in the primary care setting: connecting patients and physicians. _Alzheimer’s Dement._ 15, 321–387
(2019). MATH Google Scholar * Picón, E. et al. Does empirically derived classification of individuals with subjective cognitive complaints predict dementia. _Brain Sci._ 9, 314 (2019).
Article PubMed PubMed Central MATH Google Scholar * Afzal, S., Maqsood, M., Nazir, F., Khan, U. & Song, O. A data augmentation-based framework to handle class imbalance problem for
Alzheimer’s stage detection. _IEEE Access._ 7, 115528–115539 (2019). Article MATH Google Scholar * Schlaggar, C. N. L., Rosario, O. L. D., Morris, J. C., Ances, B. M. & Schlaggar, B.
L. Adaptation of the clinical dementia rating scale for adults with down syndrome. _J. Neurodev. Disord_. 11, 1–10 (2019). MATH Google Scholar * Umbach, G. et al. Time cells in the human
hippocampus and entorhinal cortex support episodic memory. _Proc. Natl. Acad. Sci._ 117, 28463–28474 (2020). Article ADS CAS PubMed PubMed Central MATH Google Scholar * Mehmood, S. et
al. Malignancy detection in lung and Colon histopathology images using transfer learning with Class Selective Image Processing. _IEEE Access._ 10, 25657–25668 (2022). Article MATH Google
Scholar * Taleb, N. et al. Ovary Cancer Diagnosing Empowered with Machine Learning. International Conference on Business Analytics for Technology and Security (ICBATS) 2022. 1–6. (2022). *
Nasir, M. U. et al. IoMT-Based Osteosarcoma Cancer detection in histopathology images using transfer learning empowered with Blockchain, Fog Computing, and Edge Computing. _Sensors_ 22, 5444
(2022). Article ADS PubMed PubMed Central MATH Google Scholar * Rahman, A. et al. Advance genome disorder prediction Model Empowered with Deep Learning. _IEEE Access._ 10, 70317–70328
(2022). Article Google Scholar * Rahman, A. et al. IoMT based Mitochondrial and multifactorial genetic inheritance disorder prediction using machine learning. _Comput. Intell._ 650742.
(2022). * Naseer, A. et al. Refining Parkinson’s neurological disorder identification through deep transfer learning. _Neural Comput._ 32, 839–854 (2019). Article MATH Google Scholar *
Naz, S. et al. Urdu Nastaliq recognition using convolutional recursive deep learning. _Neurocomputing_ 243, 80–87 (2017). Article MATH Google Scholar * Razzak, M. I. Malarial parasite
classification using recurrent neural network. _Int. J. Image Process._ 9, 69 (2015). MATH Google Scholar * Razzak, M. I., Naz, S. & Zaib, A. Deep learning for medical image
processing: overview, challenges, and the future. _Classif. BioApps_. 4, 323–350 (2018). Article MATH Google Scholar * Razzak, M. I. & Naz, S. Microscopic blood smear segmentation and
classification using deep contour aware CNN and extreme machine learning. IEEE Conference on Computer Vision and Pattern Recognition Workshops 49–55. (2017). * Baron, J. C. et al. In vivo
mapping of gray matter loss with Voxel-based morphometry in mild Alzheimer’s disease. _Neuroimage_ 14, 298–309 (2001). Article CAS PubMed MATH Google Scholar * Asif, S. et al. Improving
Effectiveness of Different Deep Transfer Learning-Based Models for Detecting Brain Tumors From MR Images. _IEEE Access_. 10, 34716–34730. (2022). https://doi.org/10.1109/ACCESS.2022.3153306
* Asif, S., Zhao, M., Tang, F. & Zhu, Y. An enhanced deep learning method for multi-class brain tumor classification using deep transfer learning. _Multimed. Tools Appl._ 82 (20),
31709–31736 (2023). Article Google Scholar * Khan, S. U., Rehman, M., Zhao, S., Asif & Chen, X. Hybrid-NET: a fusion of DenseNet169 and advanced machine learning classifiers for
enhanced brain tumor diagnosis. _Int. J. Imaging Syst. Technol._ 34 (1), e22975 (2024). Article Google Scholar * Asif, S., Zhao, M., Chen, X. & Zhu, Y. BMRI-NET: a deep stacked
ensemble model for multi-class brain tumor classification from MRI images. _Interdiscipl. Sci. Comput. Life Sci._ 15 (3), 499–514 (2023). Google Scholar * Zhang, F. et al. Accelerating
hyperparameter tuning in machine learning for Alzheimer’s disease with high performance computing. _Front. Artif. Intell._ 4, 798962 (2021). Article PubMed PubMed Central Google Scholar
* Hedayati, R., Khedmati, M. & Taghipour-Gorjikolaie, M. Deep feature extraction method based on ensemble of convolutional auto encoders: application to Alzheimer’s disease diagnosis.
_Biomed. Signal Process. Control_. 66, 102397 (2021). Article Google Scholar * Balaji, P., Chaurasia, M. A. & Bilfaqih, S. M. Anandhavalli Muniasamy, and Linda Elzubir Gasm Alsid.
Hybridized deep learning approach for detecting Alzheimer’s disease. _Biomedicines_ 11 (1), 149. (2023). * Plant, C. Automated detection of brain atrophy patterns based on MRI for the
prediction of Alzheimer’s disease. _Neuroimage_ 50, 162–174 (2010). Article PubMed MATH Google Scholar * Gray, R., Aljabar, P., Heckemann, R. A., Hammers, A. & Rueckert, D. Random
forest-based similarity measures for multi-modal classification of Alzheimer’s disease. _Neuroimage_ 65, 167–175 (2013). Article PubMed Google Scholar * Dahshan, E. S. A. E., Hosny, T.
& Salem, A. B. M. Hybrid intelligent techniques for MRI brain images classification. _Digit. Signal. Process. Rev. J._ 20, 433–441 (2010). Article ADS MATH Google Scholar * Ahmed, S.
Ensembles of patch-based classifiers for diagnosis of Alzheimer diseases. _IEEE Access._ 7, 73373–73383 (2019). Article Google Scholar * Hong, X. Predicting Alzheimer’s disease using
LSTM. _IEEE Access._ 7, 80893–80901 (2019). Article Google Scholar * Gupta, A., Ayhan, M. S. & Maida, A. S. Natural image bases to represent neuroimaging data. _Int. Conf. Mach.
Learn._ 28, 2024–2031 (2013). MATH Google Scholar * Suk, H., Lee, S. & Shen, D. Deep ensemble learning of sparse regression models for brain disease diagnosis. _Med. Image Anal._ 37,
101–113 (2017). Article PubMed PubMed Central MATH Google Scholar * Islam, J. & Zhang, Y. A novel deep learning based multi-class classification method for Alzheimer’s disease
detection using brain MRI data. _Lect. Notes Comput. Sci._ 10654, 213–222 (2017). Article MATH Google Scholar * Guerrero, R., Wolz, R., Rao, A. W. & Rueckert, D. Manifold population
modeling as a neuro-imaging biomarker: application to adni and adni-go. _Neuroimage_ 94, 275–286 (2014). Article CAS PubMed Google Scholar * Gorji, H. T. & Kaabouch, N. A deep
learning approach for diagnosis of mild cognitive impairment based on MRI images. _Brain Sci._ 9, 217 (2019). Article PubMed MATH Google Scholar * Hosseini-Asl, E., Keynton, R. &
El-Baz, A. Alzheimer’s disease diagnostics by adaptation of 3d convolutional network._ IEEE International Conference on Image Processing (ICIP)_ 126–130. (2016). * He, K., Zhang, X., Ren, S.
& Sun, J. Deep residual learning for image recognition. In_ Proceedings of the IEEE conference on computer vision and pattern recognition_ 770–778. (2016). * Hazarika, R. A., Kandar, D.
& Maji, A. K. An Experimental Analysis of Different deep Learning Based Models for Alzheimer’s Disease Classification Using Brain Magnetic Resonance Images. _J. King Saud Univ. Comput.
Inf. Sci._ (2021). * Shanmugam, J. V., Duraisamy, B., Simon, B. C. & Bhaskaran, P. Alzheimer’s disease classification using pre-trained deep networks. _Biomed. Signal Process. Control_.
71, 103217 (2022). Article Google Scholar * Battineni, G., Hossain, M. A., Chintalapudi, N. & Amenta, F. _Alzheimer’s Disease Classification Using Feed Forwarded Deep Neural Networks
for Brain MRI Images_. Predictive Analytics of Psychological Disorders in Healthcare. Springer, 269–283. (2022). * Sarvesh Dubey. Alzheimer’s Dataset.
https://www.kaggle.com/datasets/tourist55/alzheimers-dataset-4-class-of-images Accessed 10 June 2022. (2022). * Marcus, D. S., Fotenos, A. F., Csernansky, J. G., Morris, J. C. & Buckner,
R. L. Open access series of imaging studies: longitudinal MRI data in nondemented and demented older adults. _J. Cogn. Neurosci._ 22, 2677–2684 (2010). Article PubMed PubMed Central
Google Scholar * Krishna, S. K. & Kalluri, H. K. Deep learning and transfer learning approaches for image classification. _Int. J. Recent. Technol. Eng._ 7, 427–432 (2019). MATH Google
Scholar * MathWorks. https://www.mathworks.com/help/deeplearning/ref/trainingoptions.html Accessed 01 Aug 2022. (2022). * Sethi, M., Rani, S., Singh, A. & Mazón, J. L. V. A CAD system
for alzheimer’s disease classification using neuroimaging MRI 2D slices._ Comput. Math. Methods Med._, 2022. (2022). * Mggdadi, E., Al-Aiad, A., Al-Ayyad, M. S. & Darabseh, A. Prediction
Alzheimer’s disease from MRI images using deep learning. In_ 2021 12th International Conference on Information and Communication Systems (ICICS)_. IEEE, 120-125. (2021) * Suganthe, R. C. et
al. _Multiclass Classification of Alzheimer’s Disease Using Hybrid deep Convolutional Neural Network__ NVEO Nat. Volatiles Essent. Oils J._ 145–153. (2021). * Murugan, S. et al. DEMNET: a
deep learning model for early diagnosis of Alzheimer diseases and dementia from MR images. _Ieee Access._ 9, 90319–90329 (2021). Article MATH Google Scholar * Battineni, G., Chintalapudi,
N., Amenta, F. & Traini, E. Deep Learning Type Convolution Neural Network Architecture for Multiclass Classification of Alzheimer’s Disease._ Bioimaging_. 209–215. (2021). * Yildirim,
M. & Cinar, A. Classification of Alzheimer’s Disease MRI Images with CNN Based Hybrid Method._ Ingénierie Syst. Inf._ 25(4), 413–418. (2020). Download references FUNDING The Publication
of this study was funded by the Weill Cornell Medicine- Qatar Health Sciences Library. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Faculty of Information Technology and Computer Science,
University of Central Punjab, Lahore, 54000, Pakistan Muhammad Zahid Hussain * Department of Computer Engineering, COMSATS University Islamabad, Sahiwal Campus, Sahiwal, 57000, Pakistan
Tariq Shahzad * Department of Computer Science, Bahria University Lahore Campus, Lahore, 54000, Pakistan Shahid Mehmood * Department of Computer Science, University of Engineering and
Technology, Lahore, 54000, Pakistan Kainat Akram * Department of Software, Faculty of Artificial Intelligence and Software, Gachon University, Seongnam-si, 13120, Republic of Korea Muhammad
Adnan Khan * Abu Dhabi University, Abu Dhabi, UAE Muhammad Usman Tariq * AI Center for Precision Health, Weill Cornell Medicine-Qatar, Doha, Qatar Arfan Ahmed Authors * Muhammad Zahid
Hussain View author publications You can also search for this author inPubMed Google Scholar * Tariq Shahzad View author publications You can also search for this author inPubMed Google
Scholar * Shahid Mehmood View author publications You can also search for this author inPubMed Google Scholar * Kainat Akram View author publications You can also search for this author
inPubMed Google Scholar * Muhammad Adnan Khan View author publications You can also search for this author inPubMed Google Scholar * Muhammad Usman Tariq View author publications You can
also search for this author inPubMed Google Scholar * Arfan Ahmed View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Study conception and
design: M.Z. Hussain, T. Shahzad, S. Mehmood and K. Akram; data collection: M.Z. Hussain, T. Shahzad and K. Akram; analysis and interpretation of results: T. Shahzad, M.A. Khan, M.U. Tariq,
and A. Ahmed; draft manuscript preparation: M.Z. Hussain, M.A. Khan, M.U. Tariq and A. Ahmed. All authors reviewed the results and approved the final version of the manuscript. CORRESPONDING
AUTHORS Correspondence to Muhammad Adnan Khan or Arfan Ahmed. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. ELECTRONIC SUPPLEMENTARY MATERIAL Below is the link to the electronic
supplementary material. SUPPLEMENTARY MATERIAL 1 SUPPLEMENTARY MATERIAL 2 SUPPLEMENTARY MATERIAL 3 RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons
Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original
author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the
article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use
is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Hussain, M.Z., Shahzad, T., Mehmood, S. _et al._ A fine-tuned convolutional neural
network model for accurate Alzheimer’s disease classification. _Sci Rep_ 15, 11616 (2025). https://doi.org/10.1038/s41598-025-86635-2 Download citation * Received: 05 June 2024 * Accepted:
13 January 2025 * Published: 04 April 2025 * DOI: https://doi.org/10.1038/s41598-025-86635-2 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content:
Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative KEYWORDS *
Classification * Convolutional neural networks (CNN) * Solvers/optimizers * Alzheimer’s disease detection * Transfer learning