Play all audios:
ABSTRACT Metabolic reprogramming plays an important role in kidney cancer. We aim to investigate the causal effect of 249 metabolic biomarkers on kidney cancer from population-based data.
This study extracts data from previous genome wide association studies with large sample size. The primary endpoint is random-effect inverse variance weighted (IVW). After completing 249
times of two-sample Mendelian randomization analysis, those significant metabolites are included for further sensitivity analysis. According to a strict Bonferrion-corrected level (P <
2e-04), we only find two metabolites that are causally associated with renal cancer. They are lactate (OR:3.25, 95% CI: 1.84-5.76, P = 5.08e-05) and phospholipids to total lipids ratio in
large LDL (low density lipoprotein) (OR: 0.63, 95% CI: 0.50-0.80, P = 1.39e-04). The results are stable through all the sensitivity analysis. The results emphasize the central role of
lactate in kidney tumorigenesis and provide novel insights into possible mechanism how phospholipids could affect kidney tumorigenesis. SIMILAR CONTENT BEING VIEWED BY OTHERS GENOME-WIDE
CHARACTERIZATION OF 54 URINARY METABOLITES REVEALS MOLECULAR IMPACT OF KIDNEY FUNCTION Article Open access 02 January 2025 BLOOD METABOLIC BIOMARKERS AND COLORECTAL CANCER RISK: RESULTS FROM
LARGE PROSPECTIVE COHORT AND MENDELIAN RANDOMISATION ANALYSES Article Open access 30 April 2025 THE CAUSAL EFFECT OF SERUM AMINO ACIDS ON THE RISK OF PROSTATE CANCER: A TWO-SAMPLE MENDELIAN
RANDOMIZATION STUDY Article Open access 29 November 2024 INTRODUCTION Kidney cancer is increasingly acknowledged as a disease of metabolism1. Renal cell carcinoma (RCC), the most common
type of kidney cancer, has been identified to hold numerous gene mutations impacting basic metabolic process2,3,4. Those mutations involve in glycolysis, the tricarboxylic acid (TCA) cycle,
fatty acid oxidation, glutamine metabolism and so on. However, most of those phenomenon were based on case-control study or sample dataset research and only correlation could be
investigated5,6. Fortunately, recent publication emphasized the role of several metabolites, including lactate and fatty acid, in tumor growth7,8. They drew causal associations between
certain metabolites and kidney cancer in cell-based and animal-based models. These publication aroused our reflection that, since quantities of metabolic biomarkers existed in human blood,
which ones or even which one played critical roles in kidney cancer occurrence? As it is impractical to conduct randomized controlled studies (RCTs) to investigate the effect of a certain
metabolite concentration on kidney tumorigenesis, we decide to apply Mendelian randomization (MR) analysis to explore the causal relationship between metabolites and kidney cancer, which is
thought as the best way to exploring such a question9,10. Here, we conducted this multiple two-sample MR study to investigate the causal effect of 249 metabolic biomarkers on kidney cancer
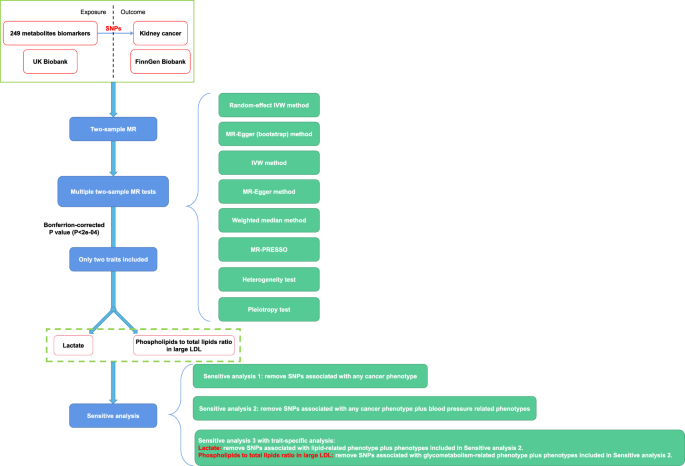
from population-based data. RESULTS Two hundred and forty-nine metabolites were tested their causal effect on kidney cancer occurrence. Finally, only two metabolic traits were considered
causal association with kidney cancer and they went on for sensitivity analysis. The study flowchart was depicted in Fig. 1. MULTIPLE TWO-SAMPLE MR ANALYSIS BETWEEN 249 METABOLIC BIOMARKERS
AND KIDNEY CANCER Totally, 249 metabolites were conducted two-sample MR analysis on kidney cancer one by one. We summarized several common metabolites’ effect on kidney cancer, including
lipidic, glycemic and fatty acid-related traits (Fig. S1). According to two-side _P_ value at 0.05 level, we discovered that lactate, sphingomyelins, phosphatidylcholines, linoleic acid,
omega-6 fatty acids, omega-3 fatty acids and polyunsaturated fatty acids had suggestive causal association with kidney cancer. All of these metabolites were positively correlated with kidney
tumorigenesis (Fig. S1). To set _P_ value at a strict Bonferrion-corrected level (_P_ < 2e–04), we only found two metabolites that were causally associated with renal cancer, among all
the 249 metabolic biomarkers (Table 1). They were lactate (OR:3.25, 95% CI: 1.84-5.76, 99.98% CI: 1.13–9.33, _P_ = 5.08e–05, Fig. 2 and Fig. S1) and phospholipids to total lipids ratio in
large LDL (low density lipoprotein) (OR: 0.63, 95% CI: 0.50–0.80, 99.98% CI: 0.41–0.98, _P_ = 1.39e-04, Fig. S2 and Fig. S3). For further details, common metabolites’ effect on renal cancer
with 99.98% CI (Bonferrion-corrected _P_ value) and 95% CI was illustrated in Fig. 2 and Fig. S1, respectively. While for phospholipid-related metabolites, two-sample MR effect was
elucidated in Fig. S2 and S3. Details about each metabolite’s effect on kidney cancer and corresponding heterogeneity and pleiotropy test results were shown in Supplementary Data 2 (MR
analysis results), Supplementary Data 3 (heterogeneity test results) and Supplementary Data 4 (pleiotropy test results). FURTHER SENSITIVITY ANALYSIS OF SIGNIFICANT METABOLIC BIOMARKERS
After multiple two-sample MR analysis between 249 metabolic biomarkers and kidney cancer, two traits were considered significant (lactate and phospholipids to total lipids ratio in large
LDL). The Steiger test results indicated a correct causal direction from the exposure variable (lactate and phospholipids to total lipids ratio in large LDL) to the outcome variable (kidney
cancer, Supplementary Data 5). Then we applied detailed sensitivity analysis. First we removed SNPs that have been reported to be significantly correlated (_P_ < 5e–08) with any cancer
phenotype (defined as sensitivity analysis 1, Fig. 3a, b). Second, we removed SNPs significantly associated with blood pressure-related phenotypes on the basis of sensitivity analysis 1
(defined as sensitivity analysis 2, Fig. 3a, b). Third, we further conducted trait-specific sensitivity analysis (defined as sensitivity analysis 3, Fig. 3a, b). For lactate biomarker, which
was thought to be involved in glycolysis, we removed SNPs significantly associated with lipid-related phenotypes on the basis of sensitivity analysis 2. While for the biomarker of
phospholipids to total lipids ratio in large LDL, which was related to lipid metabolism, we removed SNPs significantly associated with glycemic phenotypes on the basis of sensitivity
analysis 2. We described those SNPs that were utilized for sensitivity MR analysis in detail in Supplementary Data 5. Some SNPs might be left out due to their unavailability in the summary
statistics of kidney cancer phenotype. In our sensitivity analysis, we found all the results were stable (Fig. 3a, b). To be more specific, lactate was positively associated with kidney
cancer, while phospholipids to total lipids ratio in large LDL was negatively correlated with kidney cancer (Fig. 3a, b). We further performed the reverse MR analysis to validate the
positive results and avoid bidirectional causality. As a genome-wide _P_ value threshold (5e–08) excluded all the SNPs of kidney cancer variable, we used a loose P value threshold (1e–05).
The summary statistics of included SNPs were detailed in Supplementary Data 6a and Supplementary Data 6e. We did not find any causal effect of kidney cancer on lactate or phospholipids to
total lipids ratio in large LDL (beta of random-effect IVW: 0.007 for lactate and -0.002 for phospholipids to total lipids ratio in large LDL, standard error: 0.004 for lactate and 0.004 for
phospholipids to total lipids ratio in large LDL, P value: 0.138 for lactate and 0.602 for phospholipids to total lipids ratio in large LDL, Supplementary Data 6b and Supplementary Data
6f). The heterogeneity and pleiotropy effects were not observed (Supplementary Data 6c, Supplementary Data 6d, Supplementary Data 6g and Supplementary Data 6h). DISCUSSION In this study, we
conducted multiple two-sample MR analysis to investigate the causal effect of 249 metabolic biomarkers on kidney cancer risk. We did find several metabolites had suggestive causal
association with kidney cancer, which included lactate, sphingomyelins, phosphatidylcholines, linoleic acid, omega-6 fatty acids, omega-3 fatty acids and polyunsaturated fatty acids (Fig.
S1). They were common products or components necessary for renal tumor development and growth. However, after we applied a more restricted P threshold, only two traits were thought to be
significant. They were lactate (OR:3.25, 95% CI: 1.84–5.76, 99.98% CI: 1.13–9.33, _P_ = 5.08e–05, Fig. 2 and Fig. S1) and phospholipids to total lipids ratio in large LDL (OR: 0.63, 95% CI:
0.50–0.80, 99.98% CI: 0.41–0.98, _P_ = 1.39e–04, Fig. S2 and Fig. S3). These two metabolic traits played different roles in kidney tumorigenesis and the results were still stable through all
the sensitivity analysis (Fig. 3a, b). We stressed the influence of lactate in kidney tumorigenesis among all those metabolites. Actually, a recent publication revealed that perinephric
adipose tissue could release excess lactate, which promoted clear cell RCC growth, invasion and metastasis7. The study reminded us that RCC was much involved with metabolism and lactate
might serve as a key factor to facilitate the growth of kidney cancer. However, the evidence based on population of large sample size was lacking. On the other hand, numerous researches also
discovered aberrant metabolites deposition in the tissue of renal cancer or in urine4,11,12,13,14,15,16,17,18. Nevertheless, all these were just correlation analysis and a causal effect
could not be determined. Moreover, since RCC interacted with quantities of metabolites, it was difficult for us to figure out which ones were of great importance in kidney tumorigenesis
according to conventional basic and clinical studies. Luckily, recent high-quality GWAS with large sample size gave us the opportunity to investigate the causal relationship between hundreds
of metabolites and kidney cancer utilizing genetic instruments. Based on a strict P value threshold, we finally considered lactate was the most important pro-tumor factor in the development
of kidney cancer. The results could guide the direction of future researches. In addition to lactate, we also observed another metabolic biomarker, phospholipids to total lipids ratio in
large LDL, was causally negatively associated with kidney cancer and might act as a protective factor. This was a novel insight. We could see from Fig. S3, higher phospholipids to total
lipids ratio in both large and medium LDL reduced the risk of kidney cancer. How phospholipids interacted with renal cancer and whether the biomarker could be targeted as new therapy needed
further investigations. However, it has been recognized that the peroxidation of phospholipids could trigger ferroptosis, a form of programmed cell death19,20. Recent studies illustrated
that ferroptosis exerted anti-tumor effect on renal cancer21,22,23,24. From this point of view, we hypothesized that elevated phospholipids level posed a negative impact on kidney cancer
through regulating ferroptosis. Our results provided a new basis for further investigations to focus on the effect of phospholipids on kidney cancer risk. We found fatty acid might
participate in kidney tumorigenesis, although the results did not pass the strict P value threshold. In our study, we tested the effect of docosahexaenoic acid, linoleic acid, omega-3 fatty
acids, omega-6 fatty acids, polyunsaturated fatty acids, monounsaturated fatty acids and saturated fatty acids on kidney cancer. We just observed linoleic acid, omega-3 fatty acids, omega-6
fatty acids and polyunsaturated fatty acids had suggestive causal associations. These all belonged to unsaturated fatty acids, which served as necessary components of cell membrane to
maintain its fluidity. We considered it was polyunsaturated fatty acids instead of monounsaturated fatty acids or saturated fatty acids that contributed to the growth of kidney cancer. Among
those unsaturated fatty acids, omega-6 fatty acids held the strongest effect (OR: 1.58, 95% CI: 1.18-2.12). Actually, there had been one study exploring the role of fatty acid in renal
tumor growth, despite the research did not focus on the exact type of fatty acid8. The results might inspire our thinking about the causal effect of unsaturated fatty acids, particularly the
omega-6 fatty acids, on renal cancer development. The study had several strengths. Firstly, we explored the causal relationship between 249 metabolic biomarkers and kidney cancer. We
figured out that lactate was the most critical metabolite that impacted the growth of renal cancer, among all those metabolites. Secondly, we discovered the metabolic biomarker,
phospholipids to total lipids ratio in large LDL, was negatively causally associated with kidney tumorigenesis. Such findings were interesting and could facilitate further investigations.
Thirdly, we posed our reflection that polyunsaturated fatty acids rather than monounsaturated fatty acids or saturated fatty acids had the potential to induce RCC. Among those
polyunsaturated fatty acids, omega-6 fatty acids attracted the most attention due to its suggestive effect size, although not significant. The study also had limitations. Albeit we enrolled
249 metabolic biomarkers in our analysis, which covered most common metabolites, there were still several biomarkers not available in our study, for instance betaine and acetylcholine, as a
result of lacking in powerful GWAS. Moreover, we performed sensitivity analysis to excluded SNPs associated with potential confounders as possible as we could. There also existed several
unknown confounders that we might not exclude, which could violate the independence assumption. But substantially large sample size was included in our study, with strict
Bonferrion-corrected P threshold for multiple tests and several sensitivity analysis. We could still draw our conclusions with sufficient power. In addition, it was frustrating that FinnGen
Biobank did not provide summary statistics of kidney cancer with different clinical stages and histologies. That prevented us from exploring the causal effect of metabolites on the prognosis
of kidney cancer. Moreover, the Finnish population is considered a bottleneck population and genetically differentiated from other European populations, so our results should be interpreted
cautiously from this point if view. In conclusion, we identified lactate, among all 249 metabolites, that was causally associated with kidney cancer growth. Moreover, we discovered the
metabolic biomarker, phospholipids to total lipids ratio in large LDL, might serve as a protector to reduce kidney cancer risk. The results emphasized the central role of lactate in kidney
tumorigenesis and provided novel insights into possible mechanism how phospholipids could affect kidney tumorigenesis. We were looking forward to subsequent experimental verification
emphasizing on the causal role of phospholipids in kidney cancer risk. METHODS This study extracted data from previous genome wide association studies (GWAS) with large sample size. All the
data were manually curated by the MRC Integrative Epidemiology Unit (IEU) at the University of Bristol, which could be accessed through the IEU OpenGWAS project25,26. Only European
population were included. METABOLIC BIOMARKERS PHENOTYPE The summary statistics data of metabolic biomarkers were derived from UK Biobank consortium, measured by Nightingale Health 2020
(https://www.ukbiobank.ac.uk/learn-more-about-uk-biobank/news/nightingale-health-and-uk-biobank-announces-major-initiative-to-analyse-half-a-million-blood-samples-to-facilitate-global-medical-research).
Nightingale Health is a health technology company that provides a blood analysis platform for population-scale research and personalized health services. Its technology for profiling
biomarkers was utilized to examine blood samples from the UK Biobank, measuring metabolic biomarkers that have been identified in recent studies as predictors of future risk for several
common chronic diseases. In total, a variety of 249 metabolites was measured in blood samples from hundreds of thousands of participants (100,000 ~ ) and analyzed by tens of millions of
single nucleotide polymorphisms (SNPs, 10,000,000 ~ ). Briefly, these metabolites contained glycemic, lipidic, amino acid-related, fatty acid-related and some other biomarkers. More details
were described in Supplementary Data 1. The summary statistics data mainly included SNPs information (chromosome, position, effect allele, reference allele and the frequency of effect
allele), their effect size on the concentration of each metabolite (beta, standard error and P value) and sample size information. KIDNEY CANCER PHENOTYPE We utilized kidney cancer data from
FinnGen Biobank with 971 cases and 217,821 controls (Supplementary Data 1). The data excluded malignant neoplasm of renal pelvic, so only RCC samples were included. Generally, FinnGen is a
large-scale academic-industry research consortium that aims to study the genetic and environmental factors underlying common chronic diseases in the Finnish population. FinnGen has
established a biobank that includes genetic data and longitudinal health records from over 500,000 participants, making it one of the largest biobanks in the world. Its biobank contains
comprehensive health data, including electronic health records, national health registers, and biobank samples from study participants. The data is collected from various sources, such as
hospitals, health centers, and registries, and is linked with genetic data obtained from biobank samples. This allows for the identification of genetic and environmental factors that
contribute to the development of common chronic diseases, including kidney cancer. STATISTICS AND REPRODUCIBILITY We conducted such an MR study to test the causal role of “exposure” (249
metabolites) in “outcome” (kidney cancer). First, significant SNPs (_P_ < 5e–08) associated with each metabolic biomarker were extracted, after linkage disequilibrium (LD) clump (r2 <
0.001, kb = 10000). Then we extracted corresponding SNPs’ data from kidney cancer phenotype. During the process, proxy was allowed with minimum LD r2 equal to 0.8. In this way, the exposure
and outcome data were harmonized and two-sample MR effect was calculated. The primary endpoint was MR causal effect calculated by random-effect inverse variance weighted (IVW) method. We
also applied the other five methods for sensitivity analysis: IVW, MR Egger (bootstrap), MR Egger, weighted median and Mendelian Randomization Pleiotropy RESidual Sum and Outlier (MR-PRESSO)
(Fig. 1). Heterogeneity and pleiotropy test were used. After completing 249 times of two-sample MR analysis, those metabolites with significant primary endpoints were included for further
sensitivity analysis. In this way, we were able to investigate the causal effect of certain metabolites on kidney cancer. All the analysis was based on R software (4.1.2). TwoSampleMR and
ieugwasr were the main packages. The r square value was calculated as: $${R}^{2}=2 * (1-MAF) * MAF * \frac{\beta }{{{{{{\rm{S}}}}}}E * \sqrt{N}}$$ In the equation, MAF and N referred to the
minor allele frequencies and sample size, while β and SE referred to the effect size and standard error of the SNP. In our first step of multiple two-sample MR analysis, Bonferrion-corrected
two-side P value was applied (P threshold equal to 0.05/249 = 2e–04) as a result of 249 tests (Fig. 1). The results were plotted as odds ratio (OR) with 99.98% confidence interval (CI),
which was equivalent to the Bonferroni-corrected type I error rate alpha=2e-04. Any metabolite passed the threshold Bonferrion-corrected _P_ value was considered significant. While
metabolites with two-side _P_ < 0.05 were thought to be suggestive. As we intended to investigate the causal effect of 249 metabolic biomarkers on kidney cancer, it was hard to remove all
the SNPs significantly associated with confounders for each metabolites. Thus, we tried to include all the possible significant metabolites in our initial analysis. Metabolites with
significant primary endpoints were included for further strict and comprehensive sensitivity analysis to validate the causal role of significant metabolites in kidney cancer risk. The
sensitivity analysis included three steps: * i. Removal of SNPs significantly associated with any cancer phenotype, so as to eliminate possible pleiotropy; * ii. Removal of SNPs
significantly associated with any cancer phenotype plus blood pressure-related phenotypes; so as to eliminate possible pleiotropy and confounders; * iii. Trait-specific sensitivity analysis
according to the actual situation. For such sensitivity analysis, OR with 95% CI was reported to validate previous results (Fig. 1). In MR analysis, assumptions of instrumental variables
were of great importance: relevance, independence and exclusion restriction. First, we extracted significant SNPs (_P_ < 5e–08) associated with each metabolic biomarker to obey the
relevance assumption. Then, sensitivity analysis to remove SNPs associated with any cancer phenotype (particularly kidney cancer) and potential confounders was applied to stick to the
independence and exclusion restriction assumption. In addition, Steiger analysis was also performed to certify the direction of causality from the exposure variable to the outcome variable.
Lastly, the reverse MR analysis was conducted to validate the positive results and avoid bidirectional causality. REPORTING SUMMARY Further information on research design is available in the
Nature Portfolio Reporting Summary linked to this article. DATA AVAILABILITY All data generated or analyzed during this study are included in this published article and its supplementary
information files (Supplementary Data 1–6). All the phenotypes could be accessed through the corresponding id number in Supplementary Data 1 from the IEU OpenGWAS project
(https://gwas.mrcieu.ac.uk/). CODE AVAILABILITY All the analysis was based on R software (4.1.2). TwoSampleMR and ieugwasr were the main packages. REFERENCES * Yong, C., Stewart, G. D. &
Frezza, C. Oncometabolites in renal cancer. _Nat. Rev. Nephrol._ 16, 156–172 (2020). Article CAS PubMed Google Scholar * Linehan, W. M. et al. The metabolic basis of kidney cancer.
_Cancer Discov._ 9, 1006–1021 (2019). Article CAS PubMed Google Scholar * Dinges, S. S. et al. Cancer metabolomic markers in urine: evidence, techniques and recommendations. _Nat. Rev.
Urol._ 16, 339–362 (2019). Article PubMed Google Scholar * Wettersten, H. I., Aboud, O. A., Lara, P. N. Jr & Weiss, R. H. Metabolic reprogramming in clear cell renal cell carcinoma.
_Nat. Rev. Nephrol._ 13, 410–419 (2017). Article CAS PubMed Google Scholar * Breeur, M. et al. Pan-cancer analysis of pre-diagnostic blood metabolite concentrations in the European
Prospective Investigation into Cancer and Nutrition. _BMC Med_ 20, 351 (2022). Article CAS PubMed PubMed Central Google Scholar * Sun, Z. et al. Construction of a lactate-related
prognostic signature for predicting prognosis, tumor microenvironment, and immune response in kidney renal clear cell carcinoma. _Front Immunol._ 13, 818984 (2022). Article CAS PubMed
PubMed Central Google Scholar * Wei, G. et al. The thermogenic activity of adjacent adipocytes fuels the progression of ccRCC and compromises anti-tumor therapeutic efficacy. _Cell Metab._
33, 2021–2039.e8 (2021) Article CAS PubMed Google Scholar * Du, W. et al. HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. _Nat. Commun._ 8, 1769
(2017). Article PubMed PubMed Central Google Scholar * Lin, L., Wang, W., Xiao, K., Guo, X. & Zhou, L. Genetically elevated bioavailable testosterone level was associated with the
occurrence of benign prostatic hyperplasia. _J. Endocrinol. Invest_ 46, 2095–2102 (2023). Article CAS PubMed Google Scholar * Lin, L., Ning, K., Xiang, L., Peng, L. & Li, X. SGLT2
inhibition and three urological cancers: Up‐to‐date results. _Diabetes Metab. Res. Rev._ 40, e3797 (2024). * Ganti, S. et al. Urinary acylcarnitines are altered in human kidney cancer. _Int
J. Cancer_ 130, 2791–2800 (2012). Article CAS PubMed Google Scholar * Nizioł, J. et al. Metabolomic study of human tissue and urine in clear cell renal carcinoma by LC-HRMS and PLS-DA.
_Anal. Bioanal. Chem._ 410, 3859–3869 (2018). Article PubMed PubMed Central Google Scholar * Falegan, O. S. et al. Urine and serum metabolomics analyses may distinguish between stages of
renal cell carcinoma. _Metabolites_ 7, 6 (2017). Article PubMed PubMed Central Google Scholar * Hakimi, A. A. et al. An integrated metabolic atlas of clear cell renal cell carcinoma.
_Cancer Cell_ 29, 104–116 (2016). Article CAS PubMed PubMed Central Google Scholar * Wettersten, H. I. et al. Grade-dependent metabolic reprogramming in kidney cancer revealed by
combined proteomics and metabolomics analysis. _Cancer Res_ 75, 2541–2552 (2015). Article CAS PubMed PubMed Central Google Scholar * Perroud, B., Ishimaru, T., Borowsky, A. D. &
Weiss, R. H. Grade-dependent proteomics characterization of kidney cancer. _Mol. Cell Proteom._ 8, 971–985 (2009). Article CAS Google Scholar * Cancer Genome Atlas Research Network.
Comprehensive molecular characterization of clear cell renal cell carcinoma. _Nature_ 499, 43–49 (2013). * Perroud, B. et al. Pathway analysis of kidney cancer using proteomics and metabolic
profiling. _Mol. Cancer_ 5, 64 (2006). Article PubMed PubMed Central Google Scholar * Ding, K. et al. Acyl-CoA synthase ACSL4: an essential target in ferroptosis and fatty acid
metabolism. _Chin. Med J. (Engl.)_ 136, 2521–2537 (2023). Article CAS PubMed Google Scholar * Cao, L. et al. Managing ferroptosis-related diseases with indirect dietary modulators of
ferroptosis. _J. Nutr. Biochem_ 120, 109427 (2023). Article CAS PubMed Google Scholar * Hao, J. et al. Combination treatment with FAAH inhibitors/URB597 and ferroptosis inducers
significantly decreases the growth and metastasis of renal cell carcinoma cells via the PI3K-AKT signaling pathway. _Cell Death Dis._ 14, 247 (2023). Article CAS PubMed PubMed Central
Google Scholar * Li, Y. et al. Energy-stress-mediated AMPK activation promotes GPX4-dependent ferroptosis through the JAK2/STAT3/P53 axis in renal cancer. _Oxid. Med. Cell Longev._ 2022,
2353115 (2022). Article PubMed PubMed Central Google Scholar * Yangyun, W. et al. Everolimus accelerates Erastin and RSL3-induced ferroptosis in renal cell carcinoma. _Gene_ 809, 145992
(2022). Article PubMed Google Scholar * Lu, Y. et al. KLF2 inhibits cancer cell migration and invasion by regulating ferroptosis through GPX4 in clear cell renal cell carcinoma. _Cancer
Lett._ 522, 1–13 (2021). Article CAS PubMed Google Scholar * Hemani, G. et al. The MR-base platform supports systematic causal inference across the human phenome. _Elife_ 7, e34408
(2018). Article PubMed PubMed Central Google Scholar * Ben, E. et al. The MRC IEU OpenGWAS data infrastructure. _bioRxiv_ 10, 244293v1 (2020). Google Scholar Download references
ACKNOWLEDGEMENTS The work was supported by the National Natural Science Foundation of China (82303659), Natural Science Foundation of Sichuan Province (2022NSFSC1526), China Postdoctoral
Science Foundation (2022M722260) and China Primary Health Care Foundation, Urological Oncology Research Fund (016). We express our thanks to those people who have contributed to the IEU GWAS
database project and the MRC Integrative Epidemiology Unit (IEU) at the University of Bristol. Sincere thanks also go to the many GWAS consortia who have made the GWAS data that they
generated publicly available, and many members of the IEU who have contributed to curating these data. AUTHOR INFORMATION Author notes * These authors contributed equally: Lede Lin, Yaxiong
Tang, Kang Ning. AUTHORS AND AFFILIATIONS * Department of Urology and Institute of Urology, West China Hospital, Sichuan University, Chengdu, Sichuan, China Lede Lin, Yaxiong Tang, Xiang Li
& Xu Hu * Department of Head and Neck Surgery, Sun Yat-sen University Cancer Center, Guangzhou, Guangdong, China Kang Ning * State Key Laboratory of Oncology in South China, Guangdong
Provincial Clinical Research Center for Cancer, Sun Yat-sen University Cancer Center, Guangzhou, Guangdong, China Kang Ning Authors * Lede Lin View author publications You can also search
for this author inPubMed Google Scholar * Yaxiong Tang View author publications You can also search for this author inPubMed Google Scholar * Kang Ning View author publications You can also
search for this author inPubMed Google Scholar * Xiang Li View author publications You can also search for this author inPubMed Google Scholar * Xu Hu View author publications You can also
search for this author inPubMed Google Scholar CONTRIBUTIONS Conception and design of study: L.L., Y.T. and K.N. Acquisition of data: L.L., Y.T. and K.N. Data analysis and/or interpretation:
L.L., Y.T. and K.N. Drafting of manuscript and/or critical revision: L.L. and Y.T. Approval of final version of manuscript: L.L., Y.T., K.N., X.L. and X.H. CORRESPONDING AUTHORS
Correspondence to Xiang Li or Xu Hu. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Communications Biology_ thanks
the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Joao Valente. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral
with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION DESCRIPTION OF ADDITIONAL SUPPLEMENTARY MATERIALS
SUPPLEMENTARY DATA 1 SUPPLEMENTARY DATA 2 SUPPLEMENTARY DATA 3 SUPPLEMENTARY DATA 4 SUPPLEMENTARY DATA 5 SUPPLEMENTARY DATA 6 REPORTING SUMMARY RIGHTS AND PERMISSIONS OPEN ACCESS This
article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as
you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party
material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s
Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Lin, L., Tang, Y., Ning, K. _et al._
Investigating the causal associations between metabolic biomarkers and the risk of kidney cancer. _Commun Biol_ 7, 398 (2024). https://doi.org/10.1038/s42003-024-06114-8 Download citation *
Received: 09 January 2024 * Accepted: 26 March 2024 * Published: 01 April 2024 * DOI: https://doi.org/10.1038/s42003-024-06114-8 SHARE THIS ARTICLE Anyone you share the following link with
will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt
content-sharing initiative