Play all audios:
ABSTRACT The pinewood nematode, _Bursaphelenchus xylophilus_, recognized as a worldwide major forest pest, is a migratory endoparasitic nematode with capacity to feed on pine tissues and
also on fungi colonizing the trees. _Bursaphelenchus mucronatus_, the closest related species, differs from _B. xylophilus_ on its pathogenicity, making this nematode a good candidate for
comparative analyses. Secretome profiles of _B. xylophilus_ and _B. mucronatus_ were obtained and proteomic differences were evaluated by quantitative SWATH-MS. From the 681 proteins
initially identified, 422 were quantified and compared between _B. xylophilus_ and _B. mucronatus_ secretomes and from these, 243 proteins were found differentially regulated: 158 and 85
proteins were increased in _B. xylophilus_ and _B. mucronatus_ secretomes, respectively. While increased proteins in _B. xylophilus_ secretome revealed a strong enrichment in proteins with
peptidase activity, the increased proteins in _B. mucronatus_ secretome were mainly related to oxidative stress responses. The changes in peptidases were evaluated at the transcription level
by RT-qPCR, revealing a correlation between the mRNA levels of four cysteine peptidases with secretion levels. The analysis presented expands our knowledge about molecular basis of _B.
xylophilus_ and _B. mucronatu_s hosts interaction and supports the hypothesis of a key role of secreted peptidases in _B. xylophilus_ pathogenicity. SIMILAR CONTENT BEING VIEWED BY OTHERS
INTEGRATION OF TRANSCRIPTOMIC AND PROTEOMIC DATA UNCOVERS THE NUTRITIONAL REQUIREMENT OF THE IMMATURE DEVELOPMENT OF AN ENDOPARASITOID WASP Article Open access 22 May 2025 A PIPELINE
CONTRIBUTES TO EFFICIENT IDENTIFICATION OF SALIVARY PROTEINS IN SHORT-HEADED PLANTHOPPER, _EPEURYSA NAWAII_ Article Open access 14 March 2024 DIVISION OF FUNCTIONAL ROLES FOR TERMITE GUT
PROTISTS REVEALED BY SINGLE-CELL TRANSCRIPTOMES Article 08 June 2020 INTRODUCTION The pinewood nematode (PWN), _Bursaphelenchus xylophilus_, an European Plant Protection Organization A2
pest1, is an invasive species responsible for the development of the pine wilt disease (PWD) and is recognized worldwide as a major forest pest. Although it is considered a native species of
North America, American conifers are tolerant or resistant to PWN and, consequently, this nematode has caused little damage in this region2,3. At the beginning of the 20th century, PWN was
introduced in Japan and became responsible for massive mortality of native pine trees (_Pinus densiflora, P. thunbergii_ and _P. luchuensis_) and then spread to China, Korea and Taiwan3. In
Europe, it was first detected in continental Portugal in maritime pine, _P. pinaster_4 and more recently in _P. nigra_5. It has also been found in Madeira Island6 and Spain7,8 associated
with _P. pinaster_, representing an increasing threat to European conifer forests. Prevention of _B. xylophilus_ spread is particularly difficult as this nematode is vectored by bark beetles
mainly belonging to the genus _Monochamus_9. The international ecological and economic impact caused by the PWN highlight the need for further investigations on PWN pathogenic mechanisms
which are still not clear. _Bursaphelenchus xylophilus_ is a migratory endoparasitic nematode with the capacity to feed on pine tissues and also on fungi colonizing the tree. Nematodes feed
on parenchyma cells and migrate in xylem tissues, spreading throughout the tree. This causes cell destruction, leading to wilting symptoms that results in the tree death within a few months.
In the last few years, progress has been made in understanding the nature of proteins used by the PWN to successfully invade and feed on trees. Much of this progress has been supported by
large-scale expressed sequence tag (EST) transcriptomic projects undertaken for _B. xylophilus_10,11,12 and also using comparative analyses for _B. xylophilus_ and the closest related
species, _B. mucronatus_13,14. _Bursaphelenchus mucronatus_ is distributed throughout the Northern Hemisphere and is a prevalent species in Central and North Europe15. Although very similar
in morphological and ecological characteristics, _B. xylophilus_ and _B. mucronatus_ are different in pathogenicity. _Bursaphelenchus mucronatus_ was described to have weak pathogenicity and
able to kill only trees exposed to severe stress16. The higher pathogenicity of _B. xylophilus_ has been associated to its higher competitive potential and invasiveness. The higher
reproductive ability results in a rapid population growth rate which allows it to colonise new habitats more easily than _B. mucronatus_17. Also migration and pine cell destruction abilities
are important factors affecting these two species different pathogenicity and recent studies showed that the number and area of dead epithelial cells in pine cuttings inoculated with _B.
mucronatus_ were smaller than in those inoculated with _B. xylophilus_, suggesting that the attacking ability of _B. mucronatus_ is weaker than that of _B. xylophilus_18. Thus, these two
species are usually studied together for comparative analyses. A draft genome sequences of _B. xylophilus_ were also reported in 201119. The availability of all these comprehensive data sets
accelerated the postgenomic studies on PWD and a large-scale proteomic study has been conducted to better understand the pathogenicity of _B. xylophilus_20. This study presented a complete
profile of the _B. xylophilus_ secretome, however, no secretome data for _B. mucronatus_ was yet available. In the present study, SWATH-MS was used to determine changes in protein amounts
between _B. xylophilus_ and _B. mucronatu_s secretions, bringing new insights into the molecular basis of these nematodes interaction with their hosts and PWN pathogenicity. RESULTS
TRANSCRIPTOMIC PROFILES The entire reads set obtained for _B. mucronatus_ transcriptome and used for the final assembly was submitted to the EMBL-EBI European Nucleotide Archive (ENA), under
the study accession number PRJEB14884. A total of 465,256 raw pyrosequencing reads of a mean length of 337 bp were obtained for _B. mucronatus_. These were assembled in 8,822 contigs with a
mean length of 766 bp. A total of 9,231 translated amino acid sequences were deduced from contigs sequences. Annotation of the contigs resulted in 5,547 peptides associated to InterPro
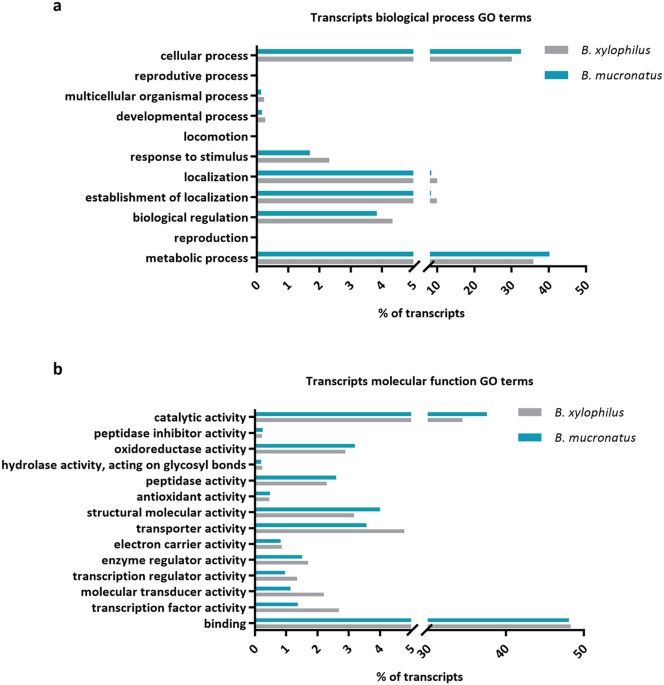
protein families or functional domains and 4,067 peptides assigned to gene ontology (GO) terms. Gene ontology analysis of _B. mucronatus_ and _B. xylophilus_ (Bioproject PRJNA192936)
transcriptomes revealed that both nematodes have a similar composition, with a higher percentage of transcripts associated with cellular and metabolic processes in biological process GO
category (Fig. 1a) and binding and catalytic activity in molecular function GO category (Fig. 1b). Analysis of higher levels of molecular function GO terms revealed that both nematodes also
have similar composition on transcripts putatively related to pathogenicity such as peptidase activity and hydrolase activity, acting on glycosyl bonds (Fig. 1b). PROTEOMIC PROFILES _General
description and global results._ From information-dependent acquisition (IDA) experiments, secretome profiles of _B. xylophilus_ (BxPE) and _B. mucronatus_ (BmPE) were obtained, using
either an annotated _B. xylophilus_ protein database derived from genome data (BioProject PRJEA64437)19 or using a combined database derived from the transcriptomic data of _B. xylophilus_
and _B. mucronatus_. A total of 520 proteins were identified in the three experimental conditions BxPE, BmPE and PE (negative control) using the genomic database (Supplementary Table S1 and
Fig. S1), while a total of 681 proteins were identified in the three conditions using the transcriptomic derived database (Supplementary Table S2), with an overlap of 50% between the two
databases (Supplementary Table S3 and Fig. S2). In BxPE condition a higher number of proteins were identified compared to the BmPE condition and, as expected, fewer proteins were identified
in PE (Fig. 2). Comparative functional analysis of _B. xylophilus_ and _B. mucronatus_ secretome profiles showed that secreted proteins have a similar GO distribution composition, with a
higher percentage of proteins associated with cellular and metabolic processes in biological process GO category (Fig. 3a) and binding and catalytic activity in molecular function GO
category (Fig. 3b). At higher levels of molecular function GO terms, only small differences were noted between both secretomes in the percentage of proteins putatively related to
pathogenicity, such as peptidase activity and hydrolase activity, acting on glycosyl bonds (Fig. 3b). _Quantitative analysis and identification of differentially secreted proteins._ From the
sequential windowed acquisition of all theoretical mass spectra (SWATH-MS) analysis, 446 proteins were quantified, considering proteins with at least one confidence peptide (with a FDR <
0.01%) in at least three out of the six biological replicates per conditions (Supplementary Table S4). According to the normal distribution of the logarithmized quantitative data,
statistical analysis was performed by multiple Student _t_-tests for each pair of conditions and proteins with P-values ≥ 0.05 in all the three comparisons were excluded. These correspond to
proteins that were at the same levels in the negative control (PE) and in the other two conditions (BxPE and BmPE). According to this evaluation, a total of 422 proteins were quantified and
compared between _B. xylophilus_ and _B. mucronatus_ secretomes (Supplementary Table S5) and from these, 243 proteins were found differentially regulated: 158 and 85 proteins were increased
in _B. xylophilus_ and _B. mucronatus_ secretomes, respectively (Fig. 4). To gain further insights into the biological differences between _B. xylophilus_ and _B. mucronatus_, functional
features of the differentially regulated proteins were characterized using GO enrichment analysis, against the entire set of quantitative data. _Bursaphelenchus xylophilus_ secretome
revealed a strong enrichment in proteins with peptidase activity and also on glycoside hydrolases activity (Table 1). The increased proteins in _B. xylophilus_ secretome associated to
peptidase activity belong to five catalytic types of peptidases and the glycoside hydrolases increased in _B. xylophilus_ secretome were mainly chitinases. Additionally, an enrichment in
proteins with peptidase inhibitor activity was also detected, one with serine and three with cysteine -type endopeptidase inhibitor activity (Table 2). On the other hand, the increased
proteins in _B. mucronatus_ secretome were mainly related to oxidative stress responses (Table 3) and from these, the proteins related to oxidoreductase activity were associated with 11
different activities (Table 4). EVALUATION OF DIFFERENTIAL TRANSCRIPT LEVEL OF CYSTEINE PEPTIDASES Cysteine and serine peptidases constituted the group of proteins with higher
representability in the increased proteins in _B. xylophilus_ secretome. In order to address whether there was a correlation between transcript level and protein level analyses, RT-qPCR was
performed for four cysteine peptidases (CP) selected from the 422 quantified proteins: CP3 and CP7, found increased in _B. xylophilus_ secretome, and CP4 and CP5, found unaltered between _B.
xylophilus_ and _B. mucronatus_ secretomes. The RT-qPCR analysis revealed that the cp3 and cp7 transcripts level was significantly (P < 0.036) higher in _B. xylophilus_ than in _B.
mucronatus_. Moreover, cp4 and cp5 transcript levels were not significantly different (P > 0.05) between both species (Fig. 5). Therefore, the patterns of changes between the two species
in transcript levels of these four genes were similar to the changes in protein levels, detected by proteomic analysis. DISCUSSION A database of _B. xylophilus_ and _B. mucronatus_
transcriptomic data was produced and used for the identification of proteins secreted by these two nematodes under pine tree extract stimulation. A general comparison of transcriptomic
profiles did not reveal notorious differences between these species and even when searching for specific groups of proteins, putatively related to nematodes pathogenicity, only small changes
were detected. This was mainly in accordance with previous studies using comparative analysis of _B. xylophilus_ and _B. mucronatus_ transcriptomic data which results indicate that the two
species have developed similar molecular mechanisms to adapt to life on pine hosts13,14. Even though, the use of this combined database in the secretome’s differential analysis allows a
higher number of identified protein comparing to those obtained using an annotated _B. xylophilus_ protein database derived from genome BioProject PRJEA6443719 and thus, constitute an
important resource for future genomic and proteomic projects on _Bursaphelenchus_ species. The identified proteins of _B. mucronatus_ secretome represent the first proteomic data on the
secretome of this species, providing new information about this nematode biology and host interaction. Furthermore, proteomic comparative and quantitative analysis with _B. xylophilus_
secretome permitted the identification of proteins detected in different levels in each secretome, reflecting a different response of these nematodes when stimulated by a pine tree extract.
No other quantitative proteomic study involving these two species has been presented before. A comparison of secretome profiles of the plant parasitic nematodes _B. xylophilus_ and
_Meloidogyne incognita_ has been previously described and the analysis of GO terms distribution indicated an expansion of peptidases and peptidase inhibitors in _B. xylophilus_ secretome20.
The similar comparative functional analysis of _B. xylophilus_ and _B. mucronatus_ secretomes, here presented, also revealed a small expansion in peptidases in _B. xylophilus_ secretome,
nevertheless it was the quantitative analysis of _B. xylophilus_ and _B. mucronatus_ secretomes that showed significant differences in protein abundances in both secretomes, pointing out
groups of proteins possibly responsible for the main differences between these two species pathogenicity. While proteins related to peptidase and glycoside hydrolase activities were detected
in higher levels in _B. xylophilus_ secretome, in _B. mucronatus_ the increased proteins were mainly related to oxidative stress responses. Peptidases are hydrolytic enzymes that cleave
internal peptide bonds within proteins and peptides. They are known to play important functions in all cellular organisms and, in nematodes, peptidases are essential not only during the
development processes such as embryogenesis and cuticle remodeling but also in the most critical moments of parasite-host interactions, such as tissue penetration, digestion of host proteins
and protection from the host immune system attack21. Peptidases can be classified according to their catalytic type and all major types of peptidases have been detected increased in _B.
xylophilus_ secretome compared to _B. mucronatus_ secretome. In other plant parasitic nematodes few reports on secreted peptidases have been presented22, however, in animal parasitic
nematodes there are many studies describing the secreted peptidases. Cysteine peptidases in animal parasitic nematodes are thought to be involved in tissue penetration, nutrition and defense
from the immune system of the host, as well as in moulting. Aspartic peptidases have been described primarily in functions related to the digestion of nutrients and metallopeptidases in
functions related to the invasion of host tissues, moulting and digestion of nutrients. The serine peptidases along with the metallopeptidases are believed to play the largest part in the
invasion of host tissues21. Identified increased peptidases in _B. xylophilus_ secretome may well have a key role in this nematode pathogenicity. On the other hand, glycoside hydrolases are
enzymes involved in carbohydrate metabolic process and are part of the known cell-wall degrading enzymes, an important group of enzymes able to break down the carbohydrates that are the
essential components of the plant and fungal cell walls. _Bursaphelenchus xylophilus_ are known to migrate in resin canals feeding on xylem parenchyma cells of pine trees but also known to
feed on fungi colonizing the trees2. In the experimental approach, the nematodes stimulated under pine tree extract were recovered from fungi cultures and it is expected that some fungi may
be present in the stimulus solution. While chitin and 1,3-beta-glucans are main components of fungi cell wall23, cellulose and the other substrates for the identified increased glycoside
hydrolases in _B. xylophilus_ secretome are components of plant cell walls. Xylem parenchyma cell walls vary among conifer species and in _Pinus_ species are mostly thin-walled and
unlignified primary walls comprising cellulose, hemicelluloses, pectins and lesser amounts of structural proteins24,25. The increased of these cell wall degrading enzymes in _B. xylophilus_
secretome compared to _B. mucronatus_ may reflect a higher capacity of this species to the feed on both plant cells and fungi colonizing trees. Feeding on xylem parenchyma cells _B.
xylophilus_ causes cell destruction, leading to the development of PWD. Fungi growing in wood tissues of diseased trees provide extra food sources and nematodes develop huge populations
whish cause the tree death within few months through damage and blocking of pine tree vascular system26. Additionally, enrichment in peptidase inhibitors were also found in increased
proteins of _B. xylophilus_ secretome and previous studies on _B. xylophilus_ secretome20 indicated that the number of secreted peptidase inhibitors in _B. xylophilus_ was significantly
greater than in other parasitic nematodes. Peptidases are known to play essential roles against pathogens in plant defence system27 and overexpression of peptidase genes in the host tree is
considered one of the most intense reactions in the case _B. xylophilus_ infection28. Therefore, these peptidase inhibitors represent an important contribute to the successful evasion of _B.
xylophilus_ from its host defence response. The increased proteins in _B. mucronatus_ secretome were mainly related to oxidative stress responses and probably play an essential role in
nematodes protection from the reactive oxygen species (ROS) accumulated inside the pine trees as a result of the host defence response. Reactive oxygen species are considered to be the first
line of defense in plants, oxidizing DNA, proteins and lipids, which causes damage to cellular organelles and inhibits cell functions in plant parasites29,30. The increased of oxidative
stress response proteins gives _B. mucronatus_ the ability to propagate and reproduce even in severe environment. The well adaptive properties of this nematode have been previously shown by
studies reporting that _B. mucronatus_ is found in declining pine trees31,32 and tends to be more active and fecund, leading to increase nematode population density inside the host tree, at
higher temperatures and drought stress33,34, which are known to enhanced ROS production in the different cellular compartments of the plant cell35,36. In the present study, antioxidant
proteins were also identified in the secretome of _B. xylophilus_. These proteins have been proved as pivotal tools in protecting _B. xylophilus_ from ROS and toxic compounds accumulated
inside the pine trees37,38. Data here presented indicate that it is quite likely that differences in _B. xylophilus_ and _B. mucronatus_ pathogenicity to pine trees are mainly related to
these peptidases, glycoside hydrolases and peptidase inhibitors increased in _B. xylophilus_ secretome. This information besides contributing to the clarification of the pathogenicity
mechanisms involved in PWD will be of great usefulness for the development of new control strategies for this important forests disease. METHODS NEMATODES Nematodes from Portuguese _B.
xylophilus_ (BxPt17AS) and _B. mucronatus_ (BmPt2) isolates, maintained in cultures of _Botrytis cinerea_ grown on Malt Extract Agar medium at 25 °C, were used. Mixed developmental nematode
stages grown during 15 days on fungal cultures were collected with distilled water using a 20 μm sieve and washed three times with sterile water. _BURSAPHELENCHUS MUCRONATUS_ TRANSCRIPTOME
SEQUENCING Total RNA was extracted from ca. 15,000 nematodes as previously described39. A fraction of 2.0 μg was used as starting material for cDNA synthesis using the MINT cDNA synthesis
kit (Evrogen), where a strategy based on SMART double stranded cDNA synthesis was applied40. cDNA was quantified by fluorescence and sequenced in a half a plate of the 454 GS-FLX Titanium
system, according to the standard manufacturer’s instructions (Roche-454 Life Sciences). Sequence reads were deposited in the EMBL-EBI European Nucleotide Archive (ENA) under the accession
number PRJEB14884. Sequence processing assembly and annotation was performed as previously described40. Prior to the assembly of sequences, the raw reads were processed in order to remove
sequences with less than 100 nucleotides and low-quality regions. The ribosomal, mitochondrial and chloroplast reads were also identified and removed from the data set. The reads were then
assembled into contigs using 454 Newbler 2.6 (Roche) with the default parameters (40 bp overlap and 90% identity). The translation frame of contigs was assessed through BLASTx searches
against Swissprot (e-value ≤ 1e–6) and the corresponding amino acid sequences translated using an in-house script. The contigs without translation were submitted to FrameDP software41 and
the remaining contigs were analysed with ESTScan42. Transcripts resulting from these two last sequence identification steps were searched using BLASTp against the non-redundant NBCI
(National Center for Biotechnology Information) database in order to translate the putative proteins. The deduced aminoacid sequences were annotated using InterProScan43 which associate each
sequence to InterPro protein families or functional domains and predicted the associated GO terms44. PREPARATION OF _BURSAPHELENCHUS XYLOPHILUS_ AND _B. MUCRONATUS_ SECRETED PROTEINS A pine
wood extract was prepared from two years old _P. pinaster_ seedlings using an adaptation of a previously described method20 and used as a stimulant for the production of secreted proteins.
Briefly, about 15 g of small wood pieces obtained from the stems were soaked in 75 mL of distilled water for 24 h at 4 °C. The collected supernatant solution was passed through a filter
paper and then centrifuged through a Vivaspin 5 kDa cutoff membrane (Sartorius Stedim). The pass-through solution containing proteins and metabolites < 5 kDa was collected and re-filtered
through a Minisart 0.2 μm cellulose acetate membrane. The obtained solution was used to stimulate the nematodes protein secretion, simulating, _in vitro_, the natural pine stimulus.
Approximately 1 × 106 nematodes of each species were soaked in 5 mL of pine extract for 16 h at 25 °C. Nematodes were then sedimented by centrifugation and the supernatants containing the
secreted proteins of _B. xylophilus_ (BxPE) and _B. mucronatus_ (BmPE) were collected and concentrated to 100 μL with a Vivaspin 5 kDa cutoff membrane. Five mL of pine extract without
nematodes were subject to the same conditions and used as control sample (PE). The sedimented nematodes were washed three times in M9 buffer, concentrated via centrifugation and used for RNA
extraction, template for reverse transcription quantitative real time PCR (RT-qPCR). Six biological replicates were performed. SAMPLE PREPARATION FOR PROTEOMIC ANALYSIS Secretomes
previously concentrated were precipitated with Trichloroacetic acid (TCA) - Acetone45. The protein pellets were ressuspended in 40 μL of SDS-Sample buffer without bromophenol blue and
glycerol46, aided by ultrasonication and denaturation at 95 °C. Two μL of each sample were used for protein quantification using the Direct Detect® infrared spectrometer (Millipore) and 60
μg of sample were used in the SWATH-MS analysis. Additionally, the six biological replicates were combined in three pools of 60 μg per condition (two biological replicates per pool) to be
used for protein identification and, the same amount of _mal_E-GFP was added. After denaturation, samples were alkylated with acrylamide and subjected to gel digestion using the short-GeLC
approach47. The entire lanes were sliced into three parts and processed in separate. Gel pieces were destained, dehydrated and re-hydrated in 70 μL of trypsin (0.01 μg/μL solution in 10 mM
ammonium bicarbonate) for 15 min, on ice. After this period, 40 μL of 10 mM ammonium bicarbonate were added and in-gel digestion was performed overnight at room temperature. After the
digestion, the formed peptides were extracted from the gel pieces and the peptides extracted from the three fractions of each biological replicate were combined into a single sample for
quantitative analysis. All the peptides were dried subjected to SPE using OMIX tips with C18 stationary phase (Agilent Technologies) as recommended by the manufacture. Eluates were dried and
ressuspended with a solution of 2% ACN and 0.1% FA containing iRT peptides (Biognosys AG) to be used as internal standards47. PROTEIN QUANTIFICATION BY SWATH-MS Samples were analysed on a
Triple TOFTM 5600 System (ABSciex®) in two phases: IDA of the pooled samples and, SWATH-MS acquisition of each individual sample. Peptides were resolved by liquid chromatography (nanoLC
Ultra 2D, Eksigent®) on a MicroLC column ChromXPTM C18CL (300 μm ID × 15 cm length, 3 μm particles, 120 Å pore size, Eksigent®) at 5 μL/min with a multistep gradient: 0–2 min linear gradient
from 5 to 10%, 2–45 min linear gradient from 10% to 30% and, 45–46 min to 35% of acetonitrile in 0.1% FA. Peptides were eluted into the mass spectrometer using an electrospray ionization
source (DuoSprayTM Source, ABSciex®) with a 50 μm internal diameter (ID) stainless steel emitter (NewObjective). Information-dependent acquisition experiments were performed for each pooled
sample. The mass spectrometer was set to scanning full spectra (350–1250 m/z) for 25 ms, followed by up to 100 MS/MS scans (100–1500 m/z from a dynamic accumulation time – minimum 30 ms for
precursor above the intensity threshold of 1000 – in order to maintain a cycle time of 3.3 s). Candidate ions with a charge state between +2 and +5 and counts above a minimum threshold of 10
counts per second were isolated for fragmentation and one MS/MS spectra was collected before adding those ions to the exclusion list for 25 seconds (mass spectrometer operated by Analyst®
TF 1.7, ABSciex®). Rolling collision was used with a collision energy spread of 5. Peptide identification and library generation were performed with Protein Pilot software (v5.1, ABSciex®)
using the following parameters: i) search against an annotated _B. xylophilus_ protein database obtained from Wormbase Parasite derived from BioProject PRJEA6443719 or a combined _B.
xylophilus_ and _B. mucronatus_ peptide database obtained from transcriptomic data; ii) acrylamide alkylated cysteines as fixed modification; iii) trypsin as digestion type. An independent
False Discovery Rate (FDR) analysis using the target-decoy approach provided with Protein Pilot software was used to assess the quality of the identifications and positive identifications
were considered when identified proteins and peptides reached a 5% local FDR48,49. Transcriptomic data is available for _B. xylophilu_s under the Bioproject PRJNA192936 and for _B.
mucronatus_ under the submitted Bioproject PRJEB14884. For SWATH-MS based experiments, the mass spectrometer was operated in a looped product ion mode50. The SWATH-MS setup was designed
specifically for the set of samples to be analysed (Supplementary Table S6), in order to adapt the SWATH windows to their complexity. A set of 60 windows of variable width was constructed
covering the precursor mass range of 350–1250 m/z. A 250 ms survey scan (350–1500 m/z) was acquired at the beginning of each cycle and SWATH MS/MS spectra were collected from 100–1500 m/z
for 50 ms resulting in a cycle time of 3.25 s. The collision energy for each window was determined according to the calculation for a charge +2 ion centered upon the window with variable
collision energy spread (CES) according with the window. A specific library of precursor masses and fragment ions was created by combining all files from the IDA experiments, and used for
subsequent SWATH processing. Libraries were obtained using Protein PilotTM software (v5.1, ABSciex®) with the same parameters as described above. Data processing was performed using SWATHTM
processing plug-in for PeakViewTM (v2.0.01, ABSciex®). After retention time adjustment using the _mal_E-GFP peptides, up to 15 peptides, with up to five fragments each, were chosen per
protein, and quantitation was attempted for all proteins from the library that were identified below 5% local FDR from ProteinPilotTM searches. Peptides’ confidence threshold was determined
based on a FDR analysis using the target-decoy approach and those that met the 1% FDR threshold in at least three of the six biological replicates were retained, and the peak areas of the
target fragment ions of those peptides were extracted across the experiments using an extracted-ion chromatogram (XIC) window of 4 min with 100 ppm XIC width. The levels of the proteins were
estimated by summing all the filtered transitions from all the filtered peptides for a given protein and normalized to the total intensity within the same experimental condition.
Correlation analysis, performed in InfernoRDN (version 1.1.5581.33355), was used to identify and excluded the less correlated replicates from the posterior analyses. Statistical analysis was
performed in MarkerViewTM (version 1.2.1.1, ABSciex®) using multiple Student _t_-test analysis for comparison between experimental groups. For statistical analysis it was used the
normalized protein levels subjected to Log10 transformation and statistical significance was considered for P-values < 0.05. Data normality was accessed by the Q-Q plots analysis
conducted in InfernoRDN (version 1.1.5581.33355). FUNCTIONAL ANNOTATION Gene ontology annotations were performed using the Blast2GO 3.3.5 software51 based on the BLAST against the
non-redundant protein database NCBI and InterPro database, using the default Blast2GO settings in each step. Gene ontology enrichment analysis of proteins increased in _B. xylophilus_ (BxPE)
and _B. mucronatus_ (BmPE) secretomes against the total number of quantified proteins were performed using Blast2GO with the statistical Fisher’s Exact Test associated and a P-value of 0.05
as cutoff. Gene ontology annotation could be assigned to three different categories: molecular function, that describe the molecular activities of gene products, cellular component that
describe where gene products are active and biological process, describing the pathways and larger processes made up of the activities of multiple gene products. MEROPS BLAST search52 was
also done to accurate the annotation of detected peptidases and peptidase inhibitors after enrichment analysis. RELATIVE TRANSCRIPT LEVEL BY RT-QPCR The relative transcript abundance of four
selected cysteine peptidases (CP3, CP4, CP5 and CP7) was assessed by RT-qPCR. Total RNA was extracted from nematodes. One hundred μL of TRIzol® Reagent (Invitrogen, Waltham, MA, USA) were
added to sedimented _B. xylophilus_ and _B. mucronatus_, vortexed for two min and subjected to three cycles of freeze and thaw in liquid nitrogen and in a 37 οC water bath. The RNA was then
purified using the Direct-zol RNA MiniPrep kit (Zymo Research Corp.), according to the manufacturer’s instructions and performing the DNase digestion in the column. Extracted RNA quality and
quantity were estimated using the spectrophotometer Nanodrop (Thermo Scientific) and the Qubit® 2.0 Fluorometer Qubit (Invitrogen) with the Qubit™ RNA assay kit (Invitrogen). For each
sample, 1 μg of extracted RNA was used for cDNA synthesis using SMARTer PCR cDNA Synthesis Kit (Clontech Laboratories Inc.) according to the manufacturer’s instructions. Synthetized cDNA was
quantified using Nanodrop and used in qPCR with SybrGreen (Applied Biosystems), according to standard protocols, in the 7500 Fast Real-Time PCR System (Applied Biosystems). The
amplification kinetics of each transcript was normalized with the amplification kinetics of the actin and 18 S genes, chosen as endogenous controls. All primers used in qPCR were designed
using the Primer Express software (Applied Biosystems) based on the _B. xylophilus_ and _B. mucronatus_ transcripts sequences, Bioprojects accession numbers PRJNA192936 and PRJEB14884,
respectively (Supplementary Table S7). qPCRs were done at 95 °C for 20 s, followed by 40 cycles of 95 °C for 3 s and 60 °C for 30 s. Melting curves analyses were performed and validation
experiments were first carried out to ensure equivalent amplification efficiency for all transcripts from both species. The RT-qPCRs were conducted for three biological repetitions, with
three technical replicates for each qPCR. Amplification efficiencies and Ct values were determined by the 7500 Fast Real-Time PCR Software v2.0.4 (Applied Biosystems) and the mean Ct values
used in the REST software53 for relative transcript level and statistically significant differences analysis using the Pair Wise Fixed Reallocation Randomisation Test©. ADDITIONAL
INFORMATION HOW TO CITE THIS ARTICLE: Cardoso, J. M. S. _et al. Bursaphelenchus xylophilus_ and _B. mucronatus_ secretomes: a comparative proteomic analysis. _Sci. Rep._ 6, 39007; doi:
10.1038/srep39007 (2016). PUBLISHER'S NOTE: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. REFERENCES * EPPO. P.
M. 7/4 (3) Bursaphelenchus xylophilus. EPPO Bull. 43, 105–118 (2013). Google Scholar * Mamiya, Y. Pathology of the Pine Wilt Disease caused by _Bursaphelenchus xylophilus_. Annu. Rev.
Phytopathol. 21, 201–220 (1983). CAS PubMed Google Scholar * Jones, J. T., Moens, M., Mota, M., Li, H. & Kikuchi, T. _Bursaphelenchus xylophilus_: Opportunities in comparative
genomics and molecular host-parasite interactions. Mol. Plant Pathol. 9, 357–368 (2008). PubMed PubMed Central Google Scholar * Mota, M. et al. First report of _Bursaphelenchus
xylophilus_ in Portugal and in Europe. Nematology 1, 727–734 (1999). Google Scholar * Inácio, M. L. et al. First detection of _Bursaphelenchus xylophilus_ associated with _Pinus nigra_ in
Portugal and in Europe. For. Pathol. 45, 235–238 (2015). Google Scholar * Fonseca, L. et al. The pinewood nematode, _Bursaphelenchus xylophilus_, in Madeira Island. Helminthologia 49,
96–103 (2012). CAS Google Scholar * Abelleira, A., Picoaga, A., Mansilla, J. P. & Aguin, O. Detection of _Bursaphelenchus xylophilus_, causal agent of pine wilt disease on _Pinus
pinaster_ in Northwestern Spain. Plant Dis. 95, 776–777 (2011). CAS PubMed Google Scholar * Robertson, L. et al. Incidence of the pinewood nematode _Bursaphelenchus xylophlius_ Steiner
& Buhrer, 1934 (Nickle, 1970) in Spain. Nematology 13, 755–757 (2011). Google Scholar * Linit, M. J. Nematode vector relationships in the Pine Wilt Disease system. J. Nematol. 20,
227–235 (1988). CAS PubMed PubMed Central Google Scholar * Espada, M. et al. Identification and characterization of parasitism genes from the pinewood nematode _Bursaphelenchus
xylophilus_ reveals a multilayered detoxification strategy. Mol. Plant Pathol. 17, 286–295 (2016). CAS PubMed Google Scholar * Tsai, I. J. et al. Transcriptional and morphological changes
in the transition from mycetophagous to phytophagous phase in the plant-parasitic nematode _Bursaphelenchus xylophilus_. Mol. Plant Pathol. 17, 77–83 (2016). CAS PubMed Google Scholar *
Kang, J. S. et al. Construction and characterization of subtractive stage-specific expressed sequence tag (EST) libraries of the pinewood nematode _Bursaphelenchus xylophilus_. Genomics 94,
70–77 (2009). CAS PubMed Google Scholar * Kikuchi, T. et al. Expressed sequence tag (EST) analysis of the pine wood nematode _Bursaphelenchus xylophilus_ and _B. mucronatus_. Mol.
Biochem. Parasitol. 155, 9–17 (2007). CAS PubMed Google Scholar * Yan, X. et al. Comparative transcriptomics of two pathogenic pinewood nematodes yields insights into parasitic adaptation
to life on pine hosts. Gene 505, 81–90 (2012). CAS PubMed Google Scholar * Pereira, F. et al. New Insights into the phylogeny and worldwide dispersion of two closely related nematode
species, _Bursaphelenchus xylophilus_ and _Bursaphelenchus mucronatus_. PLoS One 8, (2013). ADS CAS PubMed PubMed Central Google Scholar * Kanzaki, N. & Futai, K. Is
_Bursaphelenchus mucronatus_ a weak pathogen to the Japanese red pine? Nematology 8, 485–489 (2006). Google Scholar * Cheng, X. Y., Xie, P. Z., Cheng, F. X., Xu, R. M. & Xie, B. Y.
Competitive displacement of the native species _Bursaphelenchus mucronatus_ by an alien species _Bursaphelenchus xylophilus_ (Nematoda: Aphelenchida: Aphelenchoididae): A case of successful
invasion. Biol. Invasions 11, 205–213 (2009). Google Scholar * Son, J. A., Jung, C. S. & Han, H. R. Migration and attacking ability of _Bursaphelenchus mucronatus_ in Pinus thunbergii
stem cuttings. Plant Pathol. J. 32, 340–346 (2016). CAS PubMed Google Scholar * Kikuchi, T. et al. Genomic insights into the origin of parasitism in the emerging plant pathogen
_Bursaphelenchus xylophilus_. PLoS Pathog. 7 (2011). CAS PubMed PubMed Central Google Scholar * Shinya, R. et al. Secretome analysis of the Pine wood nematode _Bursaphelenchus
xylophilus_ reveals the tangled roots of parasitism and its potential for molecular mimicry. PLoS One 8, e67377 (2013). ADS CAS PubMed PubMed Central Google Scholar * Malagón, D.,
Benítez, R., Kašný, M. & Adroher, F. J. in Pa_ras_ites (ed. Erzinger, G. S. ) 61–102 (Nova Science Publishers, Inc., 2013). * Haegeman, A., Mantelin, S., Jones, J. T. & Gheysen, G.
Functional roles of effectors of plant-parasitic nematodes. Gene 492, 19–31 (2012). CAS PubMed Google Scholar * Bowman, S. M. & Free, S. J. The structure and synthesis of the fungal
cell wall. BioEssays 28, 799–808 (2006). PubMed Google Scholar * Donaldson, L. A. et al. Xylem parenchyma cell walls lack a gravitropic response in conifer compression wood. Planta 242,
1413–1424 (2015). CAS PubMed Google Scholar * Cosgrove, D. & Jarvis, M. Comparative structure and biomechanics of plant primary and secondary cell walls. Front. Plant Sci. 3, (2012).
* Futai, K. Pine wood nematode, Bursaphelenchus xylophilus. doi: 10.1146/annurev-phyto-081211-172910. (2013). CAS PubMed Google Scholar * van der Hoorn, R. A. L. & Jones, J. D. G. The
plant proteolytic machinery and its role in defence. Curr. Opin. Plant Biol. 7, 400–407 (2004). CAS PubMed Google Scholar * Hirao, T., Fukatsu, E. & Watanabe, A. Characterization of
resistance to pine wood nematode infection in _Pinus thunbergii_ using suppression subtractive hybridization. BMC Plant Biol. 12, 13 (2012). CAS PubMed PubMed Central Google Scholar *
Lamb, C. & Dixon, R. A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 251–75 (1997). CAS PubMed Google Scholar * Baker, C. J. &
Orlandi, E. W. Active Oxygen in Plant Pathogenesis. Annu. Rev. Phytopathol. 33, 299–321 (1995). Google Scholar * Polomski, J. et al. Occurrence of _Bursaphelenchus_ species in declining
_Pinus sylvestris_ in a dry Alpine valley in Switzerland. For. Pathol. 36, 110–118 (2006). Google Scholar * Oh, I. J. et al. Occurrence of _Bursaphelenchus mucronatus_ (Nematoda:
Aphelenchoididae) and the microhabitat distribution of fungi in declining pine trees in a locale in Korea. Entomol. Res. 44, 93–101 (2014). Google Scholar * Rutherford, T. a, Riga, E. &
Webster, J. M. Temperature-mediated behavioral relationships in _Bursaphelenchus xylophilus, B. mucronatus_, and their hybrids. J. Nematol. 24, 40–44 (1992). CAS PubMed PubMed Central
Google Scholar * Polomski, J. & Rigling, D. Effect of watering regime on disease development in _Pinus sylvestris_ seedlings inoculated with _Bursaphelenchus vallesianus_ and _B.
mucronatus_. Plant Dis. 94, 1055–1061 (2010). CAS PubMed Google Scholar * Cruz de Carvalho, M. H. Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant
Signal. Behav. 3, 156–65 (2008). PubMed PubMed Central Google Scholar * Suzuki, N. & Mittler, R. Reactive oxygen species and temperature stresses: A delicate balance between
signalling and destruction. Physiol. Plant. 126, 41–51 (2006). Google Scholar * Li, Z., Zhang, Q. & Zhou, X. A 2-Cys peroxiredoxin in response to oxidative stress in the pine wood
nematode, _Bursaphelenchus xylophilus_. Sci. Rep. 6, 27438 (2016). ADS CAS PubMed PubMed Central Google Scholar * Vicente, C. S. L., Ikuyo, Y., Shinya, R., Mota, M. & Hasegawa, K.
Catalases induction in high virulence pinewood nematode _Bursaphelenchus xylophilus_ under hydrogen peroxide-induced stress. PLoS One 10, 1–12 (2015). Google Scholar * Figueiredo, J. et al.
Assessment of the geographic origins of pinewood nematode isolates via single nucleotide polymorphism in effector genes. PLoS One 8, (2013). ADS PubMed PubMed Central Google Scholar *
Rocheta, M. et al. Comparative transcriptomic analysis of male and female flowers of monoecious _Quercus suber_. Front. Plant Sci. 5, 1–16 (2014). Google Scholar * Gouzy, J., Carrere, S.
& Schiex, T. FrameDP: sensitive peptide detection on noisy matured sequences. Bioinformatics 25, 670–671 (2009). CAS PubMed PubMed Central Google Scholar * Lottaz, C., Iseli, C.,
Jongeneel, C. V. & Bucher, P. Modeling sequencing errors by combining Hidden Markov models. Bioinformatics 19, II103–II112 (2003). PubMed Google Scholar * Hunter, S. et al. InterPro:
the integrative protein signature database. Nucleic Acids Res. 37, D211–D215 (2009). CAS PubMed Google Scholar * Ashburner, M. et al. Gene Ontology: tool for the unification of biology.
Nat. Genet. 25, 25–29 (2000). CAS PubMed PubMed Central Google Scholar * Anjo, S. I., Lourenço, A. S., Melo, M. N., Santa, C. & Manadas, B. in Mesenchymal Stem Cells: Methods and
Protocols (ed. Gnecchi, M. ) 521–549 (Springer: New York, 2016). doi: 10.1007/978-1-4939-3584-0_32. Google Scholar * Santa, C., Anjo, S. I. & Manadas, B. Protein precipitation of
diluted samples in SDS-containing buffer with acetone leads to higher protein recovery and reproducibility in comparison with TCA/acetone approach. Proteomics 1–5, doi:
10.1002/pmic.201600024 (2016). * Anjo, S. I., Santa, C. & Manadas, B. Short GeLC-SWATH: A fast and reliable quantitative approach for proteomic screenings. Proteomics 15, 757–762 (2015).
CAS PubMed Google Scholar * Sennels, L., Bukowski-Wills, J.-C. & Rappsilber, J. Improved results in proteomics by use of local and peptide-class specific false discovery rates. BMC
Bioinformatics 10, 179 (2009). PubMed PubMed Central Google Scholar * Tang, W. H., Shilov, I. V. & Seymour, S. L. Nonlinear fitting method for determining local false discovery rates
from decoy database searches. J. Proteome Res. 7, 3661–3667 (2008). CAS PubMed Google Scholar * Gillet, L. C. et al. Targeted data extraction of the MS/MS spectra generated by
Data-independent Acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717–O111.016717 (2012). PubMed Google Scholar * Conesa, A. et
al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005). CAS PubMed Google Scholar * Rawlings, N.
D., Waller, M., Barrett, A. J. & Bateman, A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 42, 503–509 (2014). Google Scholar *
Pfaffl, M. W. Relative expression software tool (REST(C)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, 36e–36
(2002). Google Scholar Download references ACKNOWLEDGEMENTS This research was financed by Portuguese Foundation for Science and Technology (FCT) through national funds and the co-funding by
FEDER- Fundo Europeu de Desenvolvimento Regional and COMPETE- Programa Operacional Factores de Competitividade, under the projects FCT PTDC/AGR-CFL/098916/2008 and FCT
PTDC/NEU-NMC/0205/2012, within the PT2020 Partnership Agreement and COMPETE 2020 under the projects UID/BIA/04004/2013 and UID/NEU/04539/2013, the National Mass Spectrometry Network (RNEM,
REDE/1506/REM/2005) and Instituto do Ambiente, Tecnologia e Vida, FCTUC. Joana M.S. Cardoso (SFRH/BPD/73724/2010) and Sandra I. Anjo (SFRH/BD/81495/2011) are funded by post-doctoral/doctoral
fellowships financed by QREN-POPH-Typology 4.1-co-financed by MES national funding and The European Social Fund. Luís Fonseca (SFRH/BPD/101325/2014) is funded by post-doctoral fellowship
financed by MEC national funding and by the European Social Fund through POCH (Programa Operacional Capital Humano). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Life
Sciences, CFE - Centre for Functional Ecology, University of Coimbra, Calçada Martim de Freitas, Coimbra, 3000-456, Portugal Joana M. S. Cardoso, Luís Fonseca & Isabel Abrantes * Faculty
of Sciences and Technology, University of Coimbra, Coimbra, 3030-790, Portugal Sandra I. Anjo * CNC - Center for Neuroscience and Cell Biology, University of Coimbra, Coimbra, 3004-517,
Portugal Sandra I. Anjo, Conceição Egas & Bruno Manadas Authors * Joana M. S. Cardoso View author publications You can also search for this author inPubMed Google Scholar * Sandra I.
Anjo View author publications You can also search for this author inPubMed Google Scholar * Luís Fonseca View author publications You can also search for this author inPubMed Google Scholar
* Conceição Egas View author publications You can also search for this author inPubMed Google Scholar * Bruno Manadas View author publications You can also search for this author inPubMed
Google Scholar * Isabel Abrantes View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS J.M.S.C., L.F., C.E., B.M. and I.A. conceived and designed
the experiments. J.M.S.C., S.I.A. and L.F. performed the experiments. J.M.S.C., S.I.A. and C.E. analysed the data. J.M.S.C. and S.I.A. wrote the paper. All authors discussed the results and
commented on the manuscript. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. ELECTRONIC SUPPLEMENTARY MATERIAL SUPPLEMENTARY FIGURES
SUPPLEMENTARY DATASET 1 RIGHTS AND PERMISSIONS This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article
are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to
obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ Reprints and permissions ABOUT THIS
ARTICLE CITE THIS ARTICLE Cardoso, J., Anjo, S., Fonseca, L. _et al._ _Bursaphelenchus xylophilus_ and _B. mucronatus_ secretomes: a comparative proteomic analysis. _Sci Rep_ 6, 39007
(2016). https://doi.org/10.1038/srep39007 Download citation * Received: 01 August 2016 * Accepted: 16 November 2016 * Published: 12 December 2016 * DOI: https://doi.org/10.1038/srep39007
SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to
clipboard Provided by the Springer Nature SharedIt content-sharing initiative