Play all audios:
ABSTRACT Emerging knowledge suggests that post-traumatic stress disorder (PTSD) pathophysiology is linked to the patients’ epigenetic changes, but comprehensive studies examining genome-wide
methylation have not been performed. In this study, we examined genome-wide DNA methylation in peripheral whole blood in combat veterans with and without PTSD to ascertain differentially
methylated probes. Discovery was initially made in a training sample comprising 48 male Operation Enduring Freedom (OEF)/Operation Iraqi Freedom (OIF) veterans with PTSD and 51
age/ethnicity/gender-matched combat-exposed PTSD-negative controls. Agilent whole-genome array detected ~5600 differentially methylated CpG islands (CpGI) annotated to ~2800 differently
methylated genes (DMGs). The majority (84.5%) of these CpGIs were hypermethylated in the PTSD cases. Functional analysis was performed using the DMGs encoding the promoter-bound CpGIs to
identify networks related to PTSD. The identified networks were further validated by an independent test set comprising 31 PTSD+/29 PTSD− veterans. Targeted bisulfite sequencing was also
used to confirm the methylation status of 20 DMGs shown to be highly perturbed in the training set. To improve the statistical power and mitigate the assay bias and batch effects, a union
set combining both training and test set was assayed using a different platform from Illumina. The pathways curated from this analysis confirmed 65% of the pool of pathways mined from
training and test sets. The results highlight the importance of assay methodology and use of independent samples for discovery and validation of differentially methylated genes mined from
whole blood. Nonetheless, the current study demonstrates that several important epigenetically altered networks may distinguish combat-exposed veterans with and without PTSD. SIMILAR CONTENT
BEING VIEWED BY OTHERS EPIGENETIC BIOTYPES OF POST-TRAUMATIC STRESS DISORDER IN WAR-ZONE EXPOSED VETERAN AND ACTIVE DUTY MALES Article Open access 18 December 2020 EPIGENOME-WIDE
ASSOCIATION STUDY OF POSTTRAUMATIC STRESS DISORDER IDENTIFIES NOVEL LOCI IN U.S. MILITARY VETERANS Article Open access 17 February 2022 EPIGENOME-WIDE META-ANALYSIS OF PTSD SYMPTOM SEVERITY
IN THREE MILITARY COHORTS IMPLICATES DNA METHYLATION CHANGES IN GENES INVOLVED IN IMMUNE SYSTEM AND OXIDATIVE STRESS Article 07 January 2022 INTRODUCTION Adverse life experiences alter the
epigenetic profile1, 2, 3 in a manner that is salient for pathophysiology of post-traumatic stress disorder (PTSD).4, 5, 6 Changes in methylation status of the glucocorticoid receptor gene
have been reported previously in combat veterans with PTSD.7 Methylation changes in these same genes were also observed in association with parental trauma, suggesting that such effects may
be related to heritable risk profiles.8 Consistent claims were presented by _in vivo_ studies.9, 10 Together, these discoveries drive a strong rationale for screening the epigenetic profiles
of patients’ blood to identify next-generation strategies for PTSD risk factors, diagnostics and experimental therapeutics. A growing body of cohort-based studies has linked the epigenetic
changes with PTSD development,11, 12, 13 mostly focusing on pre-determined targets such as immunity14, 15, 16 and neuroendocrinology.7, 8, 17, 18 For the present study, strict
inclusion–exclusion criteria were used19, 20 to identify a training set comprising 48 male veterans with PTSD (PTSD+) and 51 age-/ethnicity-/gender-matched controls (PTSD−). Control veterans
experienced war trauma but were negative for current and past PTSD (Supplementary Table S1). An independent test set comprising 31 PTSD+/29 PTSD− veterans was recruited using the same
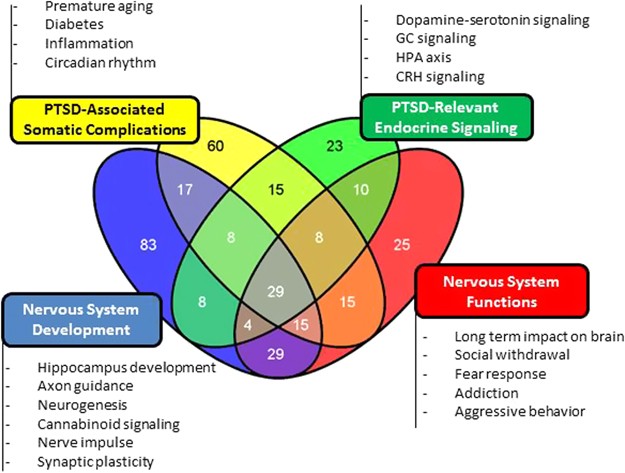
screening protocol. Enriched by the differentially methylated genes (DMGs), the epigenetically altered networks are linked to nervous systems' development and function, PTSD-associated
somatic complications and endocrine signaling. All of these networks mined from the training set were validated by the test set (Table 1). Subsequently, we consolidated the test and training
sets to develop a union set and revaluated the methylation profile using the improved sample size. The result confirmed 65% of the pathways mined from the test and training sets. Going
forward, we will consider the methylation profile from this union set as the discovery set to be confirmed in a new validation set, for which subjects are currently being recruited.
MATERIALS AND METHODS ETHICAL STATEMENT The Institutional Review Boards of the US Army Medical Research and Materiel Command, the New York University Langone Medical Center (New York, NY,
USA), the Icahn School of Medicine at Mt Sinai (New York, NY, USA) and the James J Peters Veterans Administration Medical Center (Bronx, NY, USA) approved this study. Study participants gave
written and informed consent to participate. The study was conducted in accordance with the provisions of the Helsinki Declaration. COHORT RECRUITMENT AND ANALYSIS The recruitment process
involved several steps detailed in the Supplementary Table S1 and in previous communications.19, 20 The training set of 48 PTSD+/51 PTSD− and the test set of 31 PTSD+/29 PTSD− veterans was
probed by whole-genome arrays (Agilent, Santa Clara, CA, USA) containing ~27k CpGIs. The outcome was normalized to minimize the confounding factors attributed to batch processing.21
Functional analysis was performed using those DMGs, which encoded CpGIs meeting the cutoff false discovery rate<0.1. Next, we merged the training and test sets to develop a union set
comprising 79 PTSD+/80 PTSD− veterans, which was probed by whole-genome arrays (Illumina, San Diego, CA, USA) containing 450 k probes. The outcome was corrected to minimize heterogeneous
cell populations22 and age effects, and was screened at _P_<0.05 to find DMGs. Available GEO databases are as follows: GSE76401 and GSE85399. ClueGo v2.1.2 and Ingenuity pathway analysis
were used for network construction, and pathways that we report met the cutoff of _P_<0.05. RESULTS The primary purpose of the present communication was to identify the functional
networks associated with combat-related PTSD, and thereby to provide a better understanding of PTSD pathophysiology. To meet this goal, we recruited 48 PTSD+/51 PTSD− veterans as a training
set and 31 PTSD+/29 PTSD− veterans as a test set. To increase the statistical power and to minimize any bias of the Agilent high-throughput array platform, we took two measures. First, we
constructed a union set by consolidating the training and test sets, following a recently published strategy.19, 20 Second, we retested the methylation profile, probing the union set using a
different array platform manufactured by Illumina. Furthermore, this union set retains sufficient statistical power. Taking a moderate estimate of 50% s.d.'s in probe signals and a
relatively conservative estimate for the mean difference (that is, top 1%), 76 people per group should give 95% power to detect an individual probe with a (Bonferroni-adjusted) genome-wide
significance of _P_<1.162931e−07. FUNCTIONAL ANALYSIS OF THE TRAINING SET FOUND A HOST OF PTSD-RELATED NETWORKS In the investigation of the 48 PTSD+/51 PTSD− training set, we identified
5578 differentially methylated CpGIs annotated to 3662 genes. We collectively defined the 1698 promoter-bound CpGIs and 157 additional divergent promoter regions as the _promoter_ regions
(Supplementary Figure S4A). Altogether, 4721 CpGIs annotated 2401 DMGs that displayed a log2 ratio >0.1 and were defined as hypermethylated. Conversely, 857 CpGIs (672 DMGs) displaying a
log2 ratio <0.1 were defined as hypomethylated. The remaining DMGs co-enriched by both hyper- and hypomethylated CpGIs were excluded from the subsequent functional analysis. For the
functional analysis, we used those DMGs, which encoded promoter-bound CpGIs, estimated as nearly 60% of total DMGs. Significantly enriched networks with similar functional purposes were
grouped together, resulting in four network clusters (Figure 1): nervous system functions (Figure 2a), PTSD-associated somatic complications (Figure 2b), PTSD-relevant endocrine signaling
networks (Supplementary Figure S6A) and nervous system development (Supplementary Figure S6B). TEST SET VALIDATED ALL THE NETWORKS IDENTIFIED BY THE TRAINING SET There was a significant
(_P_<0.001) overlap at the DMG level between the 48 PTSD+/51 PTSD− training set and the 31 PTSD+/29 PTSD− test set with 779 DMGs in common between the two sets assayed by the Agilent
whole-genome array. Furthermore, a significant agreement was noted at the functional level as all of the networks mined from the training set emerged significantly enriched by DMGs
identified from the test set (Table 1). UNION SET PROBED BY A DIFFERENT ARRAY PLATFORM VALIDATED A MAJORITY OF NETWORKS IDENTIFIED BY THE TRAINING AND TEST SETS The union set probed by the
Illumina array resulted in 3339 DMG, 74.4% of which encoded hypermethylated CpGIs (Supplementary Figures S4B and C). One hundred ninety-one DMGs were in common between the training set and
union set, and 107 DMGs were in common between the test set and union set (Supplementary Figure S5). There were 852 DMGs encoding promoter-bound CpGIs enriched in networks linked to
addiction, long-term impact on cerebral functions, social withdrawal, diabetes, aging, inflammation, circadian rhythm, dopamine-serotonin signaling, neurogenesis, cannabinoid signaling,
nerve impulse and synaptic plasticity. In addition, 407 DMGs in shelf and shore regions were enriched in networks associated with REM sleep, circadian rhythm, inflammation,
hypothalamic–pituitary–adrenal axis and axon guidance. Altogether, the union set confirmed 15 out of 23 networks mined from the training set and validated by test set. All of the networks
clustered under PTSD-associated somatic complications and nervous systems' development were confirmed by the training, test and union sets. METHYLATION STATUS OF SELECTED DMGS VALIDATED
BY TARGETED BISULFITE SEQUENCING Forty-two DMGs were selected from the training set based on their methylation status and their relevance to PTSD. Their methylation status was verified by
targeted bisulfite sequencing (Zymo Research, Irvine, CA, USA; Table 2).23, 24 Twenty genes out of forty-two DMGs were confirmed with the Agilent array data. Table 2 lists these genes and
their relevance to PTSD and associated comorbidities. DISCUSSION CLINICAL MEASURES WERE IN AGREEMENT WITH THE EPIGENETICALLY ALTERED NETWORKS AND DMGS Self-reported clinical measures
indicated that veterans with PTSD were concurrently experiencing higher levels of fear, social withdrawal, anxiety, hostility, depression and anger than were controls. Epigenetic
investigation of DNA extracted from whole blood revealed networks relevant to these PTSD-associated negative emotions. Greater waist size, waist-to-hip ratio and body mass index19 were found
in PTSD cases as compared with controls and are consistent with the observed pathways associated with cardiac diseases, diabetes and metabolic syndrome. PTSD-associated immune dysregulation
has been previously reported in epigenetic studies.14, 15, 19 Consistent with previous findings,14 our results found a host of innate immunity-associated genes, consisting of 60% of the
entire set of DMGs found altered in PTSD patients. In extending this knowledge, we functionally linked a majority of these genes to mobilization of phagocytic macrophages and leukocytes. In
addition, we identified epigenetically altered networks linked to learning and memory that are relevant for PTSD-associated neurocognitive impairment. Previous epidemiology studies suggested
that there was an increased risk of premature aging in PTSD.34, 35, 36 We identified two epigenetically altered networks relevant to aging. The first network is telomere management and
interaction with pathways of two mediators, wnt/β-catenin37 and p53.38 The epigenetic profile of these aging markers35 was altered in PTSD. The second network is mitochondrial dysfunction,
also epigenetically altered in PTSD veterans. Consistent with these markers of premature aging, we found evidence recently for decreased mitochondrial DNA copy numbers in PTSD veterans from
this cohort, suggesting a role for energy deprivation in PTSD that escalates the aging process.39 Premature aging40, 41 and other PTSD-associated somatic complications, such as dysregulation
of immunity,42 are known to be associated with circadian rhythm. Veterans with PTSD showed epigenetic regulation of some of the key molecular nodes responsible for setting the circadian
clock. We identified DMGs encoding CREB3 and GRIN2A, which control photoreception,43 and that are involved in signaling to entrain the circadian clock regulation by _CLOCK_ and _PER1_
genes.44 Epigenetic changes in neurogenic functional pathways were captured by the differential methylation of members of the neural helix–loop–helix family, including NEUROG1 and HES1 and
their regulators ATOH-1, Pax6 and NKX2-2.45, 46 Epigenetic perturbations of networks related to the hypothalamic–pituitary–adrenal axis functions and the synthesis of key feedback
regulators, such as corticotrophin and glucocorticoid, as well as epigenetic changes in the serotonergic and dopaminergic networks, may serve as targets for novel therapeutics for PTSD.47
STRENGTHS, LIMITATIONS AND FUTURE WORK The Diagnostic and Statistical Manual of Mental Disorders-IV diagnostic criteria48 were used to determine the PTSD status, an approach to clinical
phenotyping, which has limitations. We attempted to maximize signal detection by employing stringent selection criteria including a requirement of Clinician-Administered PTSD scale scores of
40 or greater for PTSD cases and scores less than 20 for controls.19, 20 Our array-based approach selected two platforms that ensured extensive coverage of the genome and instilled higher
confidence in the outcome. We also focused primarily on the promoter regions, as the methylation shifts near transcription start site are most likely to be associated with long-term gene
silencing.49 Given the biological heterogeneity of PTSD, our findings are limited by the sizes of our discovery, test and union sets.50 The selection of the Illumina platform was driven by
the following three factors: (i) this platform offered nearly twice the number of CpGIs to test in comparison to the Agilent platform; (ii) the significantly lower amount of input DNA
required for the Illumina assay (500 ng DNA versus 5 μg for the Agilent, assay) satisfied our need to conserve gradually decreasing DNA stocks; and (iii) the growing preference for the
Illumina assay in the epigenetics literature11, 51 was convincing for its selection. The present study recruited the largest cohort size used to date to study the PTSD pathophysiology. The
statistical analysis has moderate statistical power attributed to the sample size, which was further enhanced by the strict regulations applied by the pathway enrichment analysis. The
epigenetic contributions of many of those genes discovered have been reported as linked to PTSD via transcriptomic variations. In addition, many novel epigenetic markers linked to PTSD were
presented here. Together, this study revealed some of the key aspects of PTSD, such as its long-term health implications, which could be best explained by the epigenetic model. However, it
is challenging to draw robust mechanistic conclusions due to the non-longitudinal nature of the study; hence, there is a limited scope for making inferences about whether these epigenetic
alterations are causes of or consequences of PTSD. This study is also lacking in prospective design, gender balance and systems-wide integration. The findings are compromised further by the
fact that the array platforms are potentially unable to provide the extensive coverage typical of deep sequencing. On the basis of these findings, future work should focus on those
epigenetically altered networks presented herein, which showed clinical relevance to PTSD pathophysiology. Our study presented a knowledge-driven data-mining architecture particularly useful
to identify potential biomarkers for a multifactorial disease such as PTSD. In particular, we demonstrated how to use the clinical and physical dimensions as the successful guiding cue to
mine the molecular markers linked to disease pathophysiology. This data-mining approach will be practised further in our future study that will recruit a new validation set to confirm the
results obtained from the union set serving as the better-powered discovery set. We will also recruit a cohort of female veterans to minimize gender bias. Additional data from blood counts
and magnetic resonance imaging will be included. System-wide knowledge integration will be performed to identify PTSD biomarkers with the highest efficacy.52, 53, 54, 55, 56 REFERENCES *
Uher R, Weaver IC . Epigenetic traces of childhood maltreatment in peripheral blood: a new strategy to explore gene-environment interactions. _Br J Psychiatry_ 2014; 204: 3–5. Article
Google Scholar * Tammen SA, Friso S, Choi SW . Epigenetics: the link between nature and nurture. _Mol Aspects Med_ 2013; 34: 753–764. Article CAS Google Scholar * Feil R, Fraga MF .
Epigenetics and the environment: emerging patterns and implications. _Nat Rev Genet_ 2011; 13: 97–109. Article Google Scholar * Chakraborty N, Meyerhoffa J, Gautama A, Muhiea S, Jibitua M,
De Limab TCM _et al_. Gene and stress history interplay in emergence of PTSD-like features. _Behav Brain Res_ 2015; 292: 266–277. Article Google Scholar * Spiegel D . An ingeneious study
of intergenerational transmission of the effects of PTSD. _Am J Psychiatry_ 2014; 171: 811–813. Article Google Scholar * Stein MB, Jang KL, Taylor S, Vernon PA, Livesley WJ . Genetic and
environmental influences on trauma exposure and posttraumatic stress disorder symptoms: a twin study. _Am J Psychiatry_ 2002; 159: 1675–1681. Article Google Scholar * Yehuda R, Flory JD,
Bierer LM, Henn-Haase C, Lehrner A, Desarnaud F _et al_. Lower methylation of glucocorticoid receptor gene promoter 1F in peripheral blood of veterans with posttraumatic stress disorder.
_Biol Psychiatry_ 2015; 77: 356–364. Article CAS Google Scholar * Yehuda R, Daskalakis NP, Lehrner A, Desarnaud F, Bader HN, Makotkine I _et al_. Influences of maternal and paternal PTSD
on epigenetic regulation of the glucocorticoid receptor gene in Holocaust survivor offspring. _Am J Psychiatry_ 2014; 171: 872–880. Article Google Scholar * Zovkic IB, Sweatt JD .
Epigenetic mechanisms in learned fear: implications for PTSD. _Neuropsychopharmacology_ 2013; 38: 77–93. Article CAS Google Scholar * Pizzimenti CL, Lattal KM . Epigenetics and memory:
causes, consequences and treatments for post-traumatic stress disorder and addiction. _Genes Brain Behav_ 2015; 14: 73–84. Article CAS Google Scholar * Norrholm SD, Jovanovic T, Smith AK,
Binder E, Klengel T, Conneely K _et al_. Differential genetic and epigenetic regulation of catechol-O-methyltransferase is associated with impaired fear inhibition in posttraumatic stress
disorder. _Front Behav Neurosci_ 2013; 7: 30. Article CAS Google Scholar * Uddin M, Galea S, Chang SC, Koenen KC, Goldmann E, Wildman DE _et al_. Epigenetic signatures may explain the
relationship between socioeconomic position and risk of mental illness: preliminary findings from an urban community-based sample. _Biodemogr Soc Biol_ 2013; 59: 68–84. Article Google
Scholar * Rusiecki JA, Chen L, Srikantan V, Zhang L, Yan L, Polin ML _et al_. DNA methylation in repetitive elements and post-traumatic stress disorder: a case-control study of US military
service members. _Epigenomics_ 2012; 4: 29–40. Article CAS Google Scholar * Breen MS, Maihofer AX, Glatt SJ, Tylee DS, Chandler SD, Tsuang MT _et al_. Gene networks specific for innate
immunity define post-traumatic stress disorder. _Mol Psychiatry_ 2015; 20: 1538–1545. Article CAS Google Scholar * Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R
_et al_. Epigenetic and immune function profiles associated with posttraumatic stress disorder. _Proc Natl Acad Sci USA_ 2010; 107: 9470–9475. Article CAS Google Scholar * Rusiecki JA,
Byrne C, Galdzicki Z, Srikantan V, Chen L, Poulin M _et al_. PTSD and DNA methylation in select immune function gene promoter regions: a repeated measures case-control study of U.S. military
service members. _Front Psychiatry_ 2013; 4: 56. Article CAS Google Scholar * Vukojevic V, Kolassa IT, Fastenrath M, Gschwind L, Spalek K, Milnik A _et al_. Epigenetic modification of
the glucocorticoid receptor gene is linked to traumatic memory and post-traumatic stress disorder risk in genocide survivors. _J Neurosci_ 2014; 34: 10274–10284. Article Google Scholar *
Labonte B, Azoulay N, Yerko V, Turecki G, Brunet A . Epigenetic modulation of glucocorticoid receptors in posttraumatic stress disorder. _Transl Psychiatry_ 2014; 4: e368. Article CAS
Google Scholar * Lindqvist D, Wolkowitz OM, Mellon S, Yehuda R, Flory JD, Henn-Haase C _et al_. Proinflammatory milieu in combat-related PTSD is independent of depression and early life
stress. _Brain Behav Immun_ 2014; 42: 81–88. Article Google Scholar * Yan X, Brown AD, Lazar M, Cressman VL, Henn-Haase C, Neylan TC _et al_. Spontaneous brain activity in combat related
PTSD. _Neurosci Lett_ 2013; 547: 1–5. Article CAS Google Scholar * Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE _et al_. Tackling the widespread and critical impact of
batch effects in high-throughput data. _Nat Rev Genet_ 2010; 11: 733–739. Article CAS Google Scholar * Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH _et
al_. DNA methylation arrays as surrogate measures of cell mixture distribution. _BMC Bioinformatics_ 2012; 13: 86. Article Google Scholar * Wilmot B, Fry R, Smeester L, Musser ED, Mill J,
Nigg JT . Methylomic analysis of salivary DNA in childhood ADHD identifies altered DNA methylation in VIPR2. _J Child Psychol Psychiatr_ 2016; 57: 152–160. Article Google Scholar *
Ashktorab H, Daremipouran M, Goel A, Varma S, Leavitt R, Sun X _et al_. DNA methylome profiling identifies novel methylated genes in African American patients with colorectal neoplasia.
_Epigenetics_ 2014; 9: 503–512. Article CAS Google Scholar * Krishnan V, Han MH, Mazei-Robison M, Iniguez SD, Ables JL, Vialou V _et al_. AKT signaling within the ventral tegmental area
regulates cellular and behavioral responses to stressful stimuli. _Biol Psychiatry_ 2008; 64: 691–700. Article CAS Google Scholar * Dahlhoff M, Siegmund A, Golub Y, Wolf E, Holsboer F,
Wotjak CT . AKT/GSK-3beta/beta-catenin signalling within hippocampus and amygdala reflects genetically determined differences in posttraumatic stress disorder like symptoms. _Neuroscience_
2010; 169: 1216–1226. Article CAS Google Scholar * Martinotti G, Sepede G, Brunetti M, Ricci V, Gambi F, Chillemi E _et al_. BDNF concentration and impulsiveness level in post-traumatic
stress disorder. _Psychiatr Res_ 2015; 229: 814–818. Article CAS Google Scholar * Dell'Osso L, Carmassi C, Del Debbio A, Catena Dell'Osso M, Bianchi C, da Pozzo E _et al_.
Brain-derived neurotrophic factor plasma levels in patients suffering from post-traumatic stress disorder. _Prog Neuropsychopharmacol Biol Psychiatry_ 2009; 33: 899–902. Article CAS Google
Scholar * Su S, Xiao Z, Lin Z, Qiu Y, Jin Y, Wang Z . Plasma brain-derived neurotrophic factor levels in patients suffering from post-traumatic stress disorder. _Psychiatr Res_ 2015; 229:
365–369. Article CAS Google Scholar * Lu AT, Ogdie MN, Jarvelin MR, Moilanen IK, Loo SK, McCracken JT _et al_. Association of the cannabinoid receptor gene (CNR1) with ADHD and
post-traumatic stress disorder. _Am J Med Genet B Neuropsychiatr Genet_ 2008; 147B: 1488–1494. Article CAS Google Scholar * O'Donovan A, Sun B, Cole S, Rempel H, Lenoci M, Pulliam L
_et al_. Transcriptional control of monocyte gene expression in post-traumatic stress disorder. _Dis Markers_ 2011; 30: 123–132. Article CAS Google Scholar * Rameil P, Lecine P, Ghysdael
J, Gouilleux F, Kahn-Perles B, Imbert J . IL-2 and long-term T cell activation induce physical and functional interaction between STAT5 and ETS transcription factors in human T cells.
_Oncogene_ 2000; 19: 2086–2097. Article CAS Google Scholar * Young KA, Berry ML, Mahaffey CL, Saionz JR, Hawes NL, Chang B _et al_. Fierce: a new mouse deletion of Nr2e1; violent
behaviour and ocular abnormalities are background-dependent. _Behav Brain Res_ 2002; 132: 145–158. Article CAS Google Scholar * Zhang L, Hu XZ, Li X, Li H, Smerin S, Russell D _et al_.
Telomere length - a cellular aging marker for depression and post-traumatic stress disorder. _Med Hypotheses_ 2014; 83: 182–185. Article CAS Google Scholar * Boks MP, van Mierlo HC,
Rutten BP, Radstake TR, De Witte L, Geuze E _et al_. Longitudinal changes of telomere length and epigenetic age related to traumatic stress and post-traumatic stress disorder.
_Psychoneuroendocrinology_ 2015; 51: 506–512. Article CAS Google Scholar * Lohr JB, Palmer BW, Eidt CA, Aailaboyina S, Mausbach BT, Wolkowitz OM _et al_. Is post-traumatic stress disorder
associated with premature senescence? A review of the literature. _Am J Geriatr Psychiatry_ 2015; 23: 709–725. Article Google Scholar * Zhang DY, Wang HJ, Tan YZ . Wnt/beta-catenin
signaling induces the aging of mesenchymal stem cells through the DNA damage response and the p53/p21 pathway. _PLoS ONE_ 2011; 6: e21397. Article CAS Google Scholar * Artandi SE, Attardi
LD . Pathways connecting telomeres and p53 in senescence, apoptosis, and cancer. _Biochem Biophys Res Commun_ 2005; 331: 881–890. Article CAS Google Scholar * Bersani FS, Morley C,
Lindqvist D, Epel ES, Picard M, Yehuda R . Mitochondrial DNA copy number is reduced inmale combat veterans with PTSD. _Progr Neuro-Psychopharmacol Biol Psychiatry_ 2015; 64: 17. Google
Scholar * Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP . Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian
clock. _Genes Dev_ 2006; 20: 1868–1873. Article CAS Google Scholar * Monk TH . Aging human circadian rhythms: conventional wisdom may not always be right. _J Biol Rhythms_ 2005; 20:
366–374. Article Google Scholar * Scheiermann C, Kunisaki Y, Frenette PS . Circadian control of the immune system. _Nat Rev Immunol_ 2013; 13: 190–198. Article CAS Google Scholar *
Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P . Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. _Proc Natl Acad Sci USA_ 2002;
99: 7728–7733. Article CAS Google Scholar * Ko CH, Takahashi JS . Molecular components of the mammalian circadian clock. _Hum Mol Genet_ 2006; 15 (Spec No 2): R271–R277. Article CAS
Google Scholar * Sugimori M, Nagao M, Bertrand N, Parras CM, Guillemot F, Nakafuku M . Combinatorial actions of patterning and HLH transcription factors in the spatiotemporal control of
neurogenesis and gliogenesis in the developing spinal cord. _Development_ 2007; 134: 1617–1629. Article CAS Google Scholar * Mulvaney J, Dabdoub A . Atoh1, an essential transcription
factor in neurogenesis and intestinal and inner ear development: function, regulation, and context dependency. _J Assoc Res Otolaryngol_ 2012; 13: 281–293. Article Google Scholar * Asnis
GM, Kohn SR, Henderson M, Brown NL . SSRIs versus non-SSRIs in post-traumatic stress disorder: an update with recommendations. _Drugs_ 2004; 64: 383–404. Article CAS Google Scholar *
Association AP _Diagnostic Criteria from DSM-IV_. The Association: Washington, DC, 1994. * Jones PA . Functions of DNA methylation: islands, start sites, gene bodies and beyond. _Nat Rev
Genet_ 2012; 13: 484–492. Article CAS Google Scholar * Shao L, Fan X, Cheng N, Wu L, Cheng Y . Determination of minimum training sample size for microarray-based cancer outcome
prediction-an empirical assessment. _PLoS ONE_ 2013; 8: e68579. Article CAS Google Scholar * Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, Pace TW _et al_. Childhood maltreatment
is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. _Proc Natl Acad Sci USA_ 2013; 110: 8302–8307. Article CAS Google Scholar * Gates MA, Holowka
DW, Vasterling JJ, Keane TM, Marx BP, Rosen RC . Posttraumatic stress disorder in veterans and military personnel: epidemiology, screening, and case recognition. _Psychol Services_ 2012; 9:
361–382. Article Google Scholar * Hayes JP, Vanelzakker MB, Shin LM . Emotion and cognition interactions in PTSD: a review of neurocognitive and neuroimaging studies. _Front Integr
Neurosci_ 2012; 6: 89. Article Google Scholar * Solomon Z, Helvitz H, Zerach G . Subjective age, PTSD and physical health among war veterans. _Aging Ment Health_ 2009; 13: 405–413. Article
Google Scholar * Mehta D, Binder EB . Gene x environment vulnerability factors for PTSD: the HPA-axis. _Neuropharmacology_ 2012; 62: 654–662. Article CAS Google Scholar * McDonald SD,
Calhoun PS . The diagnostic accuracy of the PTSD checklist: a critical review. _Clin Psychol Rev_ 2010; 30: 976–987. Article Google Scholar * Thakur Gunjan S, Daigle Jr Bernie J, Dean
Kelsey R, Zhang Yuanyang, Rodriguez-Fernandez Maria, Hammamieh Rasha, Yang Ruoting, Jett Marti, Palma Joseph, Petzold Linda R, Doyle III Francis J . Systems biology approach to understanding
post-traumatic stress disorder. _Mol. BioSyst_ 2015; 11 (4): 980–993. 10.1039/C4MB00404C. Article CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS The project is supported
by the USAMRMC Military Operational Medicine Research Program (MOMRP)/Defense Health Agency (DHA)/Congressional Special Interests (CSI) and MOMRP 190040. DISCLAIMER The views, opinions
and/or findings contained in this report are those of the authors and should not be construed as an official Department of the Army position, policy or decision, unless so designated by
other official documentation. AUTHOR INFORMATION Author notes * R Hammamieh, N Chakraborty, A Gautam, S Muhie and R Yang: These authors contributed equally to this work. AUTHORS AND
AFFILIATIONS * Integrative Systems Biology, US Army Center for Environmental Health Research, Frederick, MD, USA R Hammamieh, A Gautam & M Jett * USACEHR, The Geneva Foundation,
Frederick, MD, USA N Chakraborty, S Muhie, D Donohue, S-A Miller & S Srinivasan * Advanced Biomedical Computing Center, Frederick National Laboratory for Cancer Research, Frederick, MD,
USA R Yang & R Kumar * Departments of Biological Sciences and Computer Science, The University of Memphis, Memphis, TN, USA B J Daigle Jr * School of Engineering and Applied Sciences,
Harvard University, Cambridge, MA, USA Y Zhang, L Petzold & F J Doyle III * Department of Psychiatry, Steven and Alexandra Cohen Veterans Center for the Study of Posttraumatic Stress and
Traumatic Brain Injury, NYU School of Medicine, New York, NY, USA D A Amara & C Marmar * Department of Psychiatry, Mount Sinai School of Medicine, James J Peters Veterans Administration
Medical Center, Bronx, NY, USA J Flory & R Yehuda * Department of Psychiatry, University of California San Francisco, San Francisco, CA, USA O M Wolkowitz * Department of Ob/Gyn,
Reproductive Sciences, University of California, San Francisco, San Francisco, CA, USA S H Mellon * Institute for Systems Biology, Seattle, WA, USA L Hood Authors * R Hammamieh View author
publications You can also search for this author inPubMed Google Scholar * N Chakraborty View author publications You can also search for this author inPubMed Google Scholar * A Gautam View
author publications You can also search for this author inPubMed Google Scholar * S Muhie View author publications You can also search for this author inPubMed Google Scholar * R Yang View
author publications You can also search for this author inPubMed Google Scholar * D Donohue View author publications You can also search for this author inPubMed Google Scholar * R Kumar
View author publications You can also search for this author inPubMed Google Scholar * B J Daigle Jr View author publications You can also search for this author inPubMed Google Scholar * Y
Zhang View author publications You can also search for this author inPubMed Google Scholar * D A Amara View author publications You can also search for this author inPubMed Google Scholar *
S-A Miller View author publications You can also search for this author inPubMed Google Scholar * S Srinivasan View author publications You can also search for this author inPubMed Google
Scholar * J Flory View author publications You can also search for this author inPubMed Google Scholar * R Yehuda View author publications You can also search for this author inPubMed Google
Scholar * L Petzold View author publications You can also search for this author inPubMed Google Scholar * O M Wolkowitz View author publications You can also search for this author
inPubMed Google Scholar * S H Mellon View author publications You can also search for this author inPubMed Google Scholar * L Hood View author publications You can also search for this
author inPubMed Google Scholar * F J Doyle III View author publications You can also search for this author inPubMed Google Scholar * C Marmar View author publications You can also search
for this author inPubMed Google Scholar * M Jett View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to R Hammamieh.
ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no conflict of interest. ADDITIONAL INFORMATION Supplementary Information accompanies the paper on the Translational Psychiatry
website SUPPLEMENTARY INFORMATION SUPPLEMENTARY DATA (DOCX 53 KB) SUPPLEMENTARY FIGURES (PPT 842 KB) SUPPLEMENTARY TABLE S1 (XLSX 82 KB) RIGHTS AND PERMISSIONS This work is licensed under a
Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons
license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to
reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Hammamieh, R.,
Chakraborty, N., Gautam, A. _et al._ Whole-genome DNA methylation status associated with clinical PTSD measures of OIF/OEF veterans. _Transl Psychiatry_ 7, e1169 (2017).
https://doi.org/10.1038/tp.2017.129 Download citation * Received: 03 May 2017 * Accepted: 04 May 2017 * Published: 11 July 2017 * Issue Date: July 2017 * DOI:
https://doi.org/10.1038/tp.2017.129 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently
available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative