Play all audios:
ABSTRACT Interindividual variation of the _IGF2-INS-TH_ region influences risk of a variety of diseases and complex traits. Previous studies identified a haplotype (designated
_IGF2-INS-TH_*5 and tagged by allele A of _IGF2 ApaI_, allele 9 of _TH01_ and class I alleles of _INS_ VNTR) associated with low body mass index (BMI) in a cohort of UK men. We aimed here
both to study whether previous findings relating *5 with weight are replicated in a different cohort of men (East Hertfordshire) characterised in more phenotypic detail and to test the
effect of this haplotype on related subphenotypes. The PHASE program was used to identify *5 and not*5 haplotypes. A total of 490 haplotypes were derived from 131 men and 114 women, the
frequency of *5 being around 9%. Specific tests of *5 haplotype (_vs_ not*5 haplotypes) conducted included Student's _t_-test and multiple regression analyses. We observed replication
of weight effect for the *5 haplotype in men: significant associations with lower BMI (−1.81 kg/m2, _P_=0.009), lower waist circumference (−6.3 cm, _P_=0.001) and lower waist–hip ratio (−5%,
_P_<0.001). This haplotype also marks nearly two-fold lower 120 min insulin (_P_=0.004) as well as low baseline insulin (−11.02 pmol/l, _P_=0.043) and low 30 min insulin (−64.44 pmol/l,
_P_=0.072) in a glucose tolerance test. No association between *5 and these traits was found in women. Our results, taken together with other data on IGFII levels and TH activity, point to
the importance of *5 as an integrated polygenic haplotype relevant to obesity and insulin response to glucose in men. SIMILAR CONTENT BEING VIEWED BY OTHERS UNDERSTANDING THE GENETIC
ARCHITECTURE OF THE METABOLICALLY UNHEALTHY NORMAL WEIGHT AND METABOLICALLY HEALTHY OBESE PHENOTYPES IN A KOREAN POPULATION Article Open access 26 January 2021 DISSECTING THE CLINICAL
RELEVANCE OF POLYGENIC RISK SCORE FOR OBESITY—A CROSS-SECTIONAL, LONGITUDINAL ANALYSIS Article 25 June 2022 EXPLORING THE GENETIC INTERSECTION BETWEEN OBESITY-ASSOCIATED GENETIC VARIANTS AND
INSULIN SENSITIVITY INDICES Article Open access 06 May 2025 INTRODUCTION Adult obesity is a common multifactorial disorder, which causes or exacerbates many health problems including type
II diabetes, coronary heart disease, hyperlipidaemia, hypertension, certain forms of cancer, respiratory complications and osteoarthritis of large and small joints.1, 2 Body mass index
(BMI), waist circumference (WC) and waist–hip ratio (WHR) are the most frequently used indicators of obesity and higher levels significantly reduce life expectancy.2, 3 While BMI reflects
obesity in general, WC and WHR are related to central obesity, that is, excess of body fat in the abdomen.4 It is well established that body weight and composition are substantially
determined by genetic factors that are likely to be multiple and interacting, with most single variants producing only a moderate effect.5 Estimates of heritability for obesity, measured for
BMI, have been found to be between 40 and 55% in most studies, although with estimates of up to >80% in twin studies.6 Sex differences in heritability for BMI and evidence of
sex-specific effects in BMI have been reported in a study of approximately 37 000 complete twin pairs, suggesting that there may be differences between men and women in the genetic factors
that influence variation of this trait.7 Over 600 genes, markers and chromosomal regions have been associated or linked with human obesity phenotypes.8 One such region is the
_IGF2_-_INS_-_TH_ gene cluster on human chromosome 11p15. We have previously detected a polymorphic variant of _IGF2_ in humans that associates with 10% differences in circulating levels of
IGFII and is associated with adiposity in the Northwick Park Heart Study II (NPHSII), a cohort of unrelated UK Caucasian men.9 More detailed analyses of the most common microsatellite loci
tagging haplotypes of the _IGF2_-_INS_-_TH_ cluster as defined by a set of SNPs identified in _IGF2_,10 _TH01_ alleles11 and _INS_ VNTR class I subclasses12 has identified an extended
haplotype designated _IGF2_-_INS_-_TH_*5 (hereafter *5), consisting of allele A of _IGF2 ApaI_, allele 9 of _TH01_ and a subset of class I alleles of _INS_ VNTR, which is associated with
lower BMI and fat mass in the NPHSII cohort.13 _In vitro_ functional studies (references in Rodríguez _et al_13) are also consistent with any gene of the _IGF2_-_INS_-_TH_ cluster
influencing weight. However, replication of genetic association in independent samples is important.14 In addition, significant results observed on related subphenotypes of a complex disease
may provide further insight into possible mechanism.15 We present here a follow-up analysis of _IGF2_-_INS_-_TH_*5 haplotype in an independent cohort of unrelated Caucasian individuals in
which weight association and relevant subphenotypes including response to glucose tolerance test (GTT) are examined. MATERIALS AND METHODS STUDY DESIGN The identification of the *5 haplotype
was carried out as previously described.13 In brief, we used the PHASE program version 2.016 in order to determine, with a predefined level of confidence set at ≥90%, the two haplotypes
resulting from the combination of the _TH01_ microsatellite and the SNPs _IGF2 ApaI_ and _INS HphI_ for each one of the individuals analysed. We have previously shown that the resulting
haplotypes tag the most common haplotypes of 14 polymorphisms spanning this gene cluster, giving haplotypic analyses with an increased power (due to fewer missing data) and a reduced risk of
false positive findings related to low-frequency haplotypes.13 The _IGF2 ApaI_, _INS HphI_ and _TH01_ sites were genotyped as previously described (Rodríguez _et al_13 and references
therein) from a sample of 245 unrelated UK Caucasian individuals (131 men and 114 women) from the East Hertfordshire cohort (EH). These subjects were selected from among all births in the
county of Hertfordshire UK during 1911–1930, who were followed forward and found to be alive and still resident there in 1990 to 1995. All births in Hertfordshire, from 1911 onward were
reported by the attending midwife and were recorded in ledgers. Health visitors saw the babies throughout infancy, and at 1 year of age, the babies were weighed. We computerised the ledger
entries for these subjects. Using the National Health Service Central Register (NHSCR) at Southport, we traced the cohort. Twins, triplets, and singletons who died during childhood, and boys
and girls for whom data on weight at birth and at 1 year of age were missing, were excluded. Data on many boys born in 1911–1923 were not submitted for tracing because forename, necessary
for tracing, was not recorded. We could not trace girls born before 1923 because the new names on many who married could not be determined. Ledger details were insufficient to enable NHSCR
to trace 15% of the boys and 27% of the girls.17 The subset selected for detailed evaluation of obesity-related phenotypes comprised those willing to undergo an oral GTT, they did not differ
significantly from the larger group in regard to birthweight or socioeconomic status. Both genders were nearly equally represented (53.5% of men _vs_ 46.5% of women), with similar age
distributions: age ranging in men from 59 to 70.3 years (mean±SE=64.09±0.19); and in women, from 60.3 to 71.5 years (mean±SE=64.00±0.18). The 21 traits analysed were: birthweight, weight at
1 year, weight, height, BMI, WC, hip circumference, WHR, plasma triglycerides, systolic blood pressure, diastolic blood pressure, fasting plasma concentrations of proinsulin, traits related
to oral GTT (plasma concentrations of insulin and glucose at baseline, at 30 and at 120 min after an oral load of 75 g of glucose, and insulin area under the curve during the GTT), and two
homeostatic model assessment (HOMA) variables. The two HOMA variables analysed were HOMA-%_β_ and HOMA-%S. Both models allow the deduction of _β_-cell function (%_β_) and insulin sensitivity
(or resistance) (%S), and have been validated against a variety of physiological methods.18 HOMA-%_β_ and HOMA-%S were calculated from pairs of fasting glucose and insulin measurements as
previously described.19, 20 In addition, we analyse an overall variable of the metabolic syndrome, which was obtained as described in the Adult Treatment Panel III (ATP III) report.21 In
short, five risk factors for metabolic syndrome were defined as present or absent for each individual according to established thresholds (see Table 8 from NCEP Expert Panel21). The
diagnosis of metabolic syndrome was made when three or more risk factors were present in a given individual. All subjects gave informed written consent, and permission to conduct the study
was granted by the EH ethical committee. STATISTICAL ANALYSIS As previously,13 we followed an additive haplotype-based model in this study. In keeping with the approach used in haplotype
trend regression (HTR)22 and discussed more generally,23 each individual's phenotype (e.g. weight) was used twice, once for each observed (by inference) haplotype possessed. This is
valid when Hardy–Weinberg equilibrium holds.22 The effect of the *5 haplotype on each trait was tested separately for men and women. Given prior hypothesis suggesting that *5 haplotype has
an effect on weight,13 we compared, for each trait, the effect of this haplotype with the effect of the remaining haplotypes pooled together (defined here as not*5). The differences in the
mean values observed for each trait between *5 and not*5 were compared using the Student's _t_-test. One tailed test was used for replication test of *5 haplotype–BMI association.
Adjustments for covariates were also performed using multiple regression analyses. We performed adjustments for age for analyses of all variables, and adjustments for WHR for obesity-related
phenotypes. Noting a recently published claim relating the _TH01_ microsatellite with tobacco dependence24, 25 and in order to correct for possible effects of this exposure on the
phenotypes analysed, we also adjusted for smoking status in multiple regression analyses. The overall variable of metabolic syndrome (as defined by ATP III21) was crosstabulated against the
presence/absence of the *5 haplotype, and the association tested by a standard contingency table _χ_2 test (Pearson _χ_2). The following variables were log-transformed before statistical
analyses and then retransformed for table presentation: plasma triglycerides, fasting plasma concentrations of proinsulin, insulin and glucose at baseline, plasma concentrations of insulin
and glucose at 30 and at 120 min after an oral load of 75 g of glucose, insulin area under the curve during the GTT, HOMA-%_β_ and HOMA-%S. The distributions of *5 and not*5 haplotypes were
tested for normality for all phenotypes using the Kolmogorov–Smirnov goodness-of-fit test in order to check for the presence of possible outliers. The nonparametric Kruskal–Wallis test was
also computed in order to check for concordance with the results obtained by multiple regression. The statistical analyses were conducted using SPSS (Windows version 10). RESULTS Individual
markers were all in Hardy–Weinberg equilibrium (data not shown). Tables 1 and 2 show the results obtained for *5 and for not*5 haplotypes in men and women, respectively. All 21 phenotypes
were normally distributed for both groups tested (*5 and not*5) both in men and in women (_P_>0.05 in the Kolmogorov–Smirnov goodness-of-fit test). No significant differences were
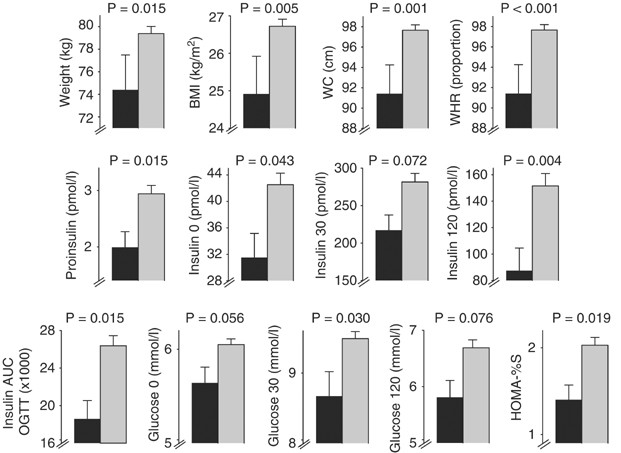
observed between *5 and not*5 in women (Table 2). In men, however, *5 associated with several traits (Table 1 and Figure 1). In accordance with previous evidence,13 haplotype *5 was found to
show a significantly lower BMI when compared with not*5 in men (_P_=0.0045). There is 1.81 unit BMI difference, whereas in the NPHSII study, *5 group averaged 0.84 units lighter than not*5
group. The corresponding mean reduction in weight observed for *5 in EH was 5 kg (2.4 kg in NPHSII). The results obtained from other obesity indicators confirm that *5 has a significant
protective effect against obesity in men. *5 shows lower WC by 6.3 cm and lower WHR by 5% (Table 1). The differences between *5 and not*5 are highly significant for BMI, WC and WHR even
after adjustments for age and smoking in multiple regression analyses (Table 1). Our results also suggest that *5 has an effect on plasma proinsulin levels, and on plasma insulin and glucose
levels both at baseline and after an oral GTT in men. A significant 1.5-fold reduction in plasma proinsulin levels was observed for *5 relative to not*5 (Table 1). However, this significant
difference depends to some extent on weight, since the _P_-value is 0.131 after adjusting for WHR in multiple regression analysis. Plasma insulin levels after the glucose load in the oral
GTT were also significantly lower for *5, the 120 min plasma insulin value remaining significant after adjustment for WHR (Table 1). The magnitudes of the differences observed in the oral
GTT between *5 and not*5 for plasma insulin levels were as follows: lower values were found for *5 in baseline levels of insulin (−11.0 pmol/l), and in plasma insulin levels after 30 min
(−64.4 pmol/l) and after 120 min (−64.1 pmol/l). These differences translate into a 1.4-fold lower insulin area under the curve during the oral GTT (_P_=0.015, Table 1). The differences in
plasma glucose levels during the oral GTT between *5 and not*5 were marginally significant: 0.41 mmol/l for baseline glucose, 0.82 mmol/l for glucose after 30 min and 0.87 mmol/l for glucose
after 120 min, _P_-values respectively 0.056, 0.030 and 0.076. In addition, the results from multiple regression analysis also showed dependence of plasma glucose levels on WHR (Table 1).
*5 associated also significantly with HOMA-%S, the magnitude of the effect observed being a reduction of 31% compared with the mean HOMA-%S value observed for not*5 haplotypes. Again, this
significant association was dependent on WHR (Table 1). The percentage of the variation explained for each trait, as measured by _R_2, varied from 1.2 to 5.9% (Table 1). Although _R_2
comparisons are difficult because they depend on the precision to which varables are measured (e.g. being more precise for plasma glucose and insulin levels than for WHR), these results
suggest that the effects of *5 on WHR, WC and plasma insulin 120 min may be larger than for the remaining variables (Table 1). No significant differences between *5 and not*5 were observed
for birthweight, weight at 1 year, height, plasma TG, systolic BP, diastolic BP or HOMA-%_β_ in men (Table 1). The nonparametric Kruskal–Wallis test confirmed all results obtained by
parametric regression (Tables 1 and 2). On the other hand, there were no significant over- or under-representations of metabolic syndrome (as defined by ATP III21) for *5 or not*5 haplotypes
either in men or in women. The percentages of metabolic syndrome observed in men for each haplotypic group were 7.8% (*5) and 8.5% (not*5); the figures observed in women were 10% (*5) and
7.8% (not*5). DISCUSSION This study describes the effects of _IGF2_-_INS_-_TH_*5 haplotype on two traits of clinical relevance: obesity and response to glucose. It both replicates evidence
of the protective effect of *5 against obesity in men and extends detail on a range of relevant subphenotypes. The observed effect of *5 on BMI is similar in both cohorts of men. The finding
presented here for the first time that WC and WHR are significantly lower for *5 suggests that the reduction in BMI conferred by this haplotype occurs mainly via reduction of visceral
obesity. The absence of association between *5 and obesity-related phenotypes in women is in accordance with previous evidence suggesting differences between both sexes in the genetic
factors influencing weight.7 The *5 haplotype seems not to be associated with metabolic syndrome (as defined by ATP III21). This however does not argue against our results, since we have
shown that *5 associates with two out of the five risk factors established by ATP III,21 and this definition of metabolic syndrome assumes the presence of at least three of such risk
factors. The replication found in this study used a smaller sample size relative to our previous work.13 It has been suggested that the effect sizes may often be smaller in replication
studies than in the initial study in which association has been reported.26 However, for this replication study, a simple dichotomy focused on the presence _vs_ absence of *5 haplotype
reduced 10-fold the need for Bonferroni correction (when considering haplotypes *1 to *10), and justifiable one-sided test represented a more powerful test. In fact, the effect size was also
larger. It could be argued that the multiple phenotypes examined would require a Bonferroni correction for 21 independent tests. However, many of the traits are interrelated (e.g. all the
weight-related traits, all the GTT related traits, and systolic and diastolic BP), which reduces the number of independent tests conducted. A more realistic Bonferroni correction of 5 would
leave most signals significant. In addition, it seems unlikely that a statistical artifact led to consistency both of significance and direction of haplotypic effects of *5 for two
independent cohorts. On the contrary, the BMI findings and other trait features, taken together with our previous findings13 support that the associations found have a biological basis and
argues against type-I error or possible biases of the sampling scheme followed (the later supported also by the fact that all 21 phenotypes were normally distributed for *5 and not*5
haplotypes). The replication found in this work has greater relevance given the high risk of false negative results expected for SNP and haplotype studies27 and the low replication rate
reported for association studies of obesity.28 The recently reported association of an individual SNP in _IGF2_ with BMI, WC and fat percentage in 538 males from 92 extended Finnish
families29 also points to an effect of _IGF2_, although the SNPs studied have not been related to our *1 to *10 haplotype classification.13 Other literature also suggests that the
_IGF2_-_INS_-_TH_ region may be important in obesity.30, 31, 32, 33, 34, 35, 36, 37, 38 The confirmation of *5 as a weight-lowering haplotype in men and our findings on related phenotypes
suggest that the _IGF2_-_INS_-_TH_ region harbours causal site(s) influencing obesity and insulin response to glucose. Haplotype association analyses _per se_ do not permit estimation either
of the physical location of a causal genetic variant or its support interval. However, the extended haplotype used, which combines multiallelic microsatellite information with SNPs, has
proved to be more powerful than the analysis of each of its single constituents, since we have not found significant associations for _IGF2 ApaI_, _INS HphI_ and _TH01_ in this data set
(data not shown). Other approaches to determination of causal SNPs (e.g. Maniatis _et al_39) may determine the presence of one or more causal sites on this haplotype. Our results, however,
permit speculation about possible functional element(s) of *5. There is evidence in the literature involving all three genes, _IGF2_, _INS_ and _TH_, in the regulation of body weight. It is
known that insulin is a very efficient inhibitor of free fatty acid mobilisation. Insulin decreases the release of free fatty acids from adipose tissue by inhibiting hormone-sensitive lipase
and stimulates triacylglycerol uptake into adipose tissue by activating lipoprotein lipase.33 Thus, a gradual decline in insulin secretion stimulates free fatty acid release from the
adipose tissue.38 Therefore, a low expressor haplotype of insulin could be a primary determinant of lower weight. Indeed, we have observed in this work lower proinsulin and lower baseline
plasma insulin levels in association with *5. However, it is also known that increase of insulin secretion in proportion to accumulated fat counteracts insulin resistance and protects
individuals from hyperglycemia.40 Therefore, it is also possible that the lower insulin levels after an oral GTT found for *5 could be secondary to the lesser insulin resistance of lean
individuals. In this case (see below), _IGF2_ could contain the haplotypic determinants of the leanness. Multiple regression results suggest that most of the insulin and glucose effects
observed for *5 are substantially dependent on WHR, a statistical pointer that the insulin and glucose associations of *5 may be secondary to its weight effects. The same applies to the
results we obtained for insulin sensitivity (or resistance) as measured by HOMA-%S. However, the fact that plasma insulin levels 120 min after the oral GTT are significantly lower for *5
after adjustment for WHR, suggests a dual role of this haplotype on both phenotypes. Longitudinal studies may resolve these possibilities. Previous evidence has related variation in the
_INS_ VNTR with control of insulin expression. However, the results obtained have been mutually inconsistent, some studies relating class I alleles with higher insulin levels,40, 41, 42 some
showing association of class III alleles with increased insulin levels,43, 44, 45 and others reporting no effect of this VNTR on insulin level variation.46, 47 The *5 haplotype contains
_INS_ VNTR class I alleles. However, it is not the only haplotype containing class I alleles (so do *1, *2, *3, *7, *9, *10) whereas these haplotypes tested individually do not show weight
effect (Rodríguez _et al_13 and this study) nor GTT effects (this study, data not shown). Indeed, our data suggest that the _INS_ VNTR may not be the potential single causal site for the
associations with weight and insulin response observed in the present study. This, however, does not exclude that the *5 haplotype harbours a causal site determining expression of insulin
and the shorter lengths of the small subclass of class I alleles associated with *5 haplotypes could be relevant.48 Complete resequencing of this haplotype together with functional studies
at the level of insulin expression may be necessary to search for such a putative causal site. Association of class III homozygotes at the _INS_ VNTR with greater birth weight has been
reported,49 although more recent studies in Hertfordshire men,50 in other UK cohorts51 and in a large Finnish birth cohort,52 did not corroborate this association. Neither did
_IGF2-INS-TH_*5 haplotype associate with birth weight or weight at 1 year in Hertfordshire men in this analysis. *5 haplotype does not represent class III _vs_ class I but is associated with
the subset of smallest class I alleles. Of additional note, *5 haplotype associated strongly with much lower 120 min insulin levels in a GTT (Table 1), whereas no major difference was
observed comparing class I and class III homozygotes (data not shown). The functional element(s) of *5, which determine the weight effect, could also reside in the _TH_ and/or _IGF2_ genes.
It is known that visceral fat is more lipolytic in response to catecholamine stimulation than is subcutaneous fat.35 Also known is that the _TH01_ microsatellite located in intron 1 of the
_TH_ gene has a role in the transcription of this gene, which encodes the rate-limiting enzyme in catecholamine biosynthesis, transcription appearing to be inhibited in proportion to the
number of repeats from 3 to 8 and with a counteracted effect for alleles above eight repeats.53 The lower visceral fat associated with *5 could reflect an increased lipolysis of visceral fat
stimulated by catecholamines via a comparatively increased expression of _TH_ conferred by the *5 constituent _TH01_ allele 9 (with a core of 9 repeats). However, it should be pointed out
that haplotype *9 (containing _TH01_ allele 9) as well as haplotypes *3, *4 and *6 (all containing allele 9.3) do not show a weight-lowering effect.13 Recent studies of IGFII suggest that
this major fetal growth factor with effects on fetal skeletal muscle growth and development, which is also expressed in adult life, may be associated with lipid metabolism and obesity. The
expression of an _Igf2_ transgene in adult skin reduces the amount of body fat,30 whereas low plasma IGFII concentrations predict weight gain and obesity.54 Homozygotes for allele A at _IGF2
ApaI_ site in the 3′ noncoding region show a mean body weight 3.3 kg lighter and show higher mean serum IGFII levels than the common (GG) homozygotes.9 It is plausible, therefore, that *5
predicts high IGFII levels and that this high expressor status is a primary protection against obesity. However, previous evidence showing that haplotype *6, which also contains _IGF2 ApaI_
A allele, does not show a weight-lowering effect,13 indicates that the _ApaI_ site either has no functional effect or has effect conditional to other haplotypic elements. In conclusion, our
haplotypic analyses confirm that *5 has a protective effect against obesity in men and the investigation, for the first time, of its effect on related phenotypes, suggests that *5 may also
be relevant to variation of insulin levels in GTT in men. _IGF2_, _INS_ and _TH_ are certainly haplotypically coupled and may conceivably be functionally (evolutionarily) coupled in their
effect(s) both on weight and on insulin response to glucose. REFERENCES * Lamon-Fava S, Wilson PW, Schaefer EJ : Impact of body mass index on coronary heart disease risk factors in men and
women. The Framingham Offspring Study. _Arterioscler Thromb Vasc Biol_ 1996; 16: 1509–1515. Article CAS Google Scholar * Kopelman PG : Obesity as a medical problem. _Nature_ 2000; 404:
635–643. Article CAS Google Scholar * Friedman JM : Obesity in the new millennium. _Nature_ 2000; 404: 632–634. Article CAS Google Scholar * Sonmez K, Akcakoyun M, Akcay A _et al_:
Which method should be used to determine the obesity, in patients with coronary artery disease? (body mass index, waist circumference or waist–hip ratio). _Int J Obes Relat Metab Disord_
2003; 27: 341–346. Article CAS Google Scholar * Barsh GS, Farooqi IS, O'Rahilly S : Genetics of body-weight regulation. _Nature_ 2000; 404: 644–651. Article CAS Google Scholar *
Feitosa MF, Borecki IB, Rich SS _et al_: Quantitative-trait loci influencing body-mass index reside on chromosomes 7 and 13: the National Heart, Lung, and Blood Institute Family Heart Study.
_Am J Hum Genet_ 2002; 70: 72–82. Article CAS Google Scholar * Schousboe K, Willemsen G, Kyvik KO _et al_: Sex differences in heritability of BMI: a comparative study of results from
twin studies in eight countries. _Twin Res_ 2003; 6: 409–421. Article Google Scholar * Perusse L, Rankinen T, Zuberi A _et al_: The human obesity gene map: the 2004 update. _Obes Res_
2005; 13: 381–490. Article CAS Google Scholar * O'Dell SD, Miller GJ, Cooper JA _et al_: Apal polymorphism in insulin-like growth factor II (IGF2) gene and weight in middle-aged
males. _Int J Obes Relat Metab Disord_ 1997; 21: 822–825. Article CAS Google Scholar * Gaunt TR, Cooper JA, Miller GJ, Day IN, O'Dell SD : Positive associations between single
nucleotide polymorphisms in the IGF2 gene region and body mass index in adult males. _Hum Mol Genet_ 2001; 10: 1491–1501. Article CAS Google Scholar * Puers C, Hammond HA, Jin L, Caskey
CT, Schumm JW : Identification of repeat sequence heterogeneity at the polymorphic short tandem repeat locus HUMTH01[AATG]n and reassignment of alleles in population analysis by using a
locus-specific allelic ladder. _Am J Hum Genet_ 1993; 53: 953–958. CAS PubMed PubMed Central Google Scholar * Gu D, O'Dell SD, Chen XH, Miller GJ, Day IN : Evidence of multiple
causal sites affecting weight in the IGF2-INS-TH region of human chromosome 11. _Hum Genet_ 2002; 110: 173–181. Article CAS Google Scholar * Rodríguez S, Gaunt TR, O'Dell SD _et al_:
Haplotypic analyses of the IGF2-INS-TH gene cluster in relation to cardiovascular risk traits. _Hum Mol Genet_ 2004; 13: 715–725. Article Google Scholar * Cardon LR, Bell JI : Association
study designs for complex diseases. _Nat Rev Genet_ 2001; 2: 91–99. Article CAS Google Scholar * Campbell H, Rudan I : Interpretation of genetic association studies in complex disease.
_Pharmacogenomics J_ 2002; 2: 349–360. Article CAS Google Scholar * Stephens M, Donnelly P : A comparison of bayesian methods for haplotype reconstruction from population genotype data.
_Am J Hum Genet_ 2003; 73: 1162–1169. Article CAS Google Scholar * Barker DJ, Winter PD, Osmond C, Margetts B, Simmonds SJ : Weight in infancy and death from ischaemic heart disease.
_Lancet_ 1989; 2: 577–580. Article CAS Google Scholar * Wallace TM, Levy JC, Matthews DR : Use and abuse of HOMA modeling. _Diabetes Care_ 2004; 27: 1487–1495. Article Google Scholar *
Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC : Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin
concentrations in man. _Diabetologia_ 1985; 28: 412–419. Article CAS Google Scholar * Levy JC, Matthews DR, Hermans MP : Correct homeostasis model assessment (HOMA) evaluation uses the
computer program. _Diabetes Care_ 1998; 21: 2191–2192. Article CAS Google Scholar * NCEP Expert Panel: Executive Summary of The Third Report of The National Cholesterol Education Program
(NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III). _JAMA_ 2001; 285: 2486–2497. Article Google Scholar * Zaykin
DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG : Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals.
_Hum Hered_ 2002; 53: 79–91. Article Google Scholar * Sasieni PD : From genotypes to genes: doubling the sample size. _Biometrics_ 1997; 53: 1253–1261. Article CAS Google Scholar *
Anney RJ, Olsson CA, Lotfi-Miri M, Patton GC, Williamson R : Nicotine dependence in a prospective population-based study of adolescents: the protective role of a functional tyrosine
hydroxylase polymorphism. _Pharmacogenetics_ 2004; 14: 73–81. Article CAS Google Scholar * Olsson C, Anney R, Forrest S _et al_: Association between dependent smoking and a polymorphism
in the tyrosine hydroxylase gene in a prospective population-based study of adolescent health. _Behav Genet_ 2004; 34: 85–91. Article Google Scholar * Colhoun HM, McKeigue PM, Davey-Smith
G : Problems of reporting genetic associations with complex outcomes. _Lancet_ 2003; 361: 865–872. Article Google Scholar * Neale BM, Sham PC : The future of association studies:
gene-based analysis and replication. _Am J Hum Genet_ 2004; 75: 353–362. Article CAS Google Scholar * Redden DT, Allison DB : Nonreplication in genetic association studies of obesity and
diabetes research. _J Nutr_ 2003; 133: 3323–3326. Article CAS Google Scholar * Suviolahti E, Soro-Paavonen A, Silander K _et al_: Variants in IGF2-IGF2AS-INS gene cluster on chromosome
11p15 are associated with obesity related traits in Finns. _Atherosclerosis Supplements_ 2004; 5: 60. Article Google Scholar * Da Costa TH, Williamson DH, Ward A _et al_: High plasma
insulin-like growth factor-II and low lipid content in transgenic mice: measurements of lipid metabolism. _J Endocrinol_ 1994; 143: 433–439. Article CAS Google Scholar * Nezer C, Moreau
L, Brouwers B _et al_: An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. _Nat Genet_ 1999; 21: 155–156. Article CAS Google Scholar *
Jeon JT, Carlborg O, Tornsten A _et al_: A paternally expressed QTL affecting skeletal and cardiac muscle mass in pigs maps to the IGF2 locus. _Nat Genet_ 1999; 21: 157–158. Article CAS
Google Scholar * Jequier E, Tappy L : Regulation of body weight in humans. _Physiol Rev_ 1999; 79: 451–480. Article CAS Google Scholar * Jones BK, Levorse J, Tilghman SM : Deletion of a
nuclease-sensitive region between the Igf2 and H19 genes leads to Igf2 misregulation and increased adiposity. _Hum Mol Genet_ 2001; 10: 807–814. Article CAS Google Scholar * Camp HS, Ren
D, Leff T : Adipogenesis and fat-cell function in obesity and diabetes. _Trends Mol Med_ 2002; 8: 442–447. Article CAS Google Scholar * Rice T, Chagnon YC, Perusse L _et al_: A genomewide
linkage scan for abdominal subcutaneous and visceral fat in black and white families: The HERITAGE Family Study. _Diabetes_ 2002; 51: 848–855. Article CAS Google Scholar * Sandhu MS,
Gibson JM, Heald AH, Dunger DB, Wareham NJ : Low circulating IGF-II concentrations predict weight gain and obesity in humans. _Diabetes_ 2003; 52: 1403–1408. Article CAS Google Scholar *
van Dijk G, de Vries K, Benthem L, Nyakas C, Buwalda B, Scheurink AJ : Neuroendocrinology of insulin resistance: metabolic and endocrine aspects of adiposity. _Eur J Pharmacol_ 2003; 480:
31–42. Article CAS Google Scholar * Maniatis N, Collins A, Gibson J, Zhang W, Tapper W, Morton NE : Positional cloning by linkage disequilibrium. _Am J Hum Genet_ 2004; 74: 846–855.
Article CAS Google Scholar * Le Stunff C, Fallin D, Schork NJ, Bougneres P : The insulin gene VNTR is associated with fasting insulin levels and development of juvenile obesity. _Nat
Genet_ 2000; 26: 444–446. Article CAS Google Scholar * Bazaes RA, Petry CJ, Ong KK, Avila A, Dunger DB, Mericq MV : Insulin gene VNTR genotype is associated with insulin sensitivity and
secretion in infancy. _Clin Endocrinol (Oxf)_ 2003; 59: 599–603. Article CAS Google Scholar * Dos Santos C, Fallin D, Le Stunff C, LeFur S, Bougneres P : INS VNTR is a QTL for the insulin
response to oral glucose in obese children. _Physiol Genomics_ 2004; 16: 309–313. Article CAS Google Scholar * Vafiadis P, Bennett ST, Todd JA _et al_: Insulin expression in human thymus
is modulated by INS VNTR alleles at the IDDM2 locus. _Nat Genet_ 1997; 15: 289–292. Article CAS Google Scholar * Pugliese A, Zeller M, Fernandez Jr A _et al_: The insulin gene is
transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. _Nat Genet_ 1997; 15: 293–297.
Article CAS Google Scholar * Ong KK, Petry CJ, Barratt BJ _et al_: Maternal-fetal interactions and birth order influence insulin variable number of tandem repeats allele class
associations with head size at birth and childhood weight gain. _Diabetes_ 2004; 53: 1128–1133. Article CAS Google Scholar * Ahmed S, Bennett ST, Huxtable SJ, Todd JA, Matthews DR, Gough
SC : INS VNTR allelic variation and dynamic insulin secretion in healthy adult non-diabetic Caucasian subjects. _Diabetes Med_ 1999; 16: 910–917. Article CAS Google Scholar * Hansen SK,
Gjesing AP, Rasmussen SK _et al_: Large-scale studies of the HphI insulin gene variable-number-of-tandem-repeats polymorphism in relation to Type 2 diabetes mellitus and insulin release.
_Diabetologia_ 2004; 47: 1079–1087. Article CAS Google Scholar * O'Dell SD, Bujac SR, Miller GJ, Day IN : Associations of IGF2 ApaI RFLP and INS VNTR class I allele size with
obesity. _Eur J Hum Genet_ 1999; 7: 821–827. Article CAS Google Scholar * Dunger DB, Ong KK, Huxtable SJ _et al_: Association of the INS VNTR with size at birth. ALSPAC Study Team. Avon
Longitudinal Study of Pregnancy and Childhood. _Nat Genet_ 1998; 19: 98–100. Article CAS Google Scholar * Ong KK, Phillips DI, Fall C _et al_: The insulin gene VNTR, type 2 diabetes and
birth weight. _Nat Genet_ 1999; 21: 262–263. Article CAS Google Scholar * Mitchell SM, Hattersley AT, Knight B _et al_: Lack of support for a role of the insulin gene variable number of
tandem repeats minisatellite (INS-VNTR) locus in fetal growth or type 2 diabetes-related intermediate traits in United Kingdom populations. _J Clin Endocrinol Metab_ 2004; 89: 310–317.
Article CAS Google Scholar * Bennett AJ, Sovio U, Ruokonen A _et al_: Variation at the insulin gene VNTR (variable number tandem repeat) polymorphism and early growth: studies in a large
Finnish Birth Cohort. _Diabetes_ 2004; 53: 2126–2131. Article CAS Google Scholar * Albanese V, Biguet NF, Kiefer H, Bayard E, Mallet J, Meloni R : Quantitative effects on gene silencing
by allelic variation at a tetranucleotide microsatellite. _Hum Mol Genet_ 2001; 10: 1785–1792. Article CAS Google Scholar * Sandhu MS, Gibson JM, Heald AH, Dunger DB, Wareham NJ : Low
circulating IGF-II concentrations predict weight gain and obesity in humans. _Diabetes_ 2003; 52: 1403–1408. Article CAS Google Scholar Download references ACKNOWLEDGEMENTS The UK MRC is
thanked for support. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Human Genetics Division, University of Southampton, School of Medicine, Duthie Building (MP 808), Southampton General
Hospital, Tremona Road, Southampton, UK Santiago Rodríguez, Tom R Gaunt, Xiao-he Chen & Ian N M Day * Fetal Origins of Adult Disease Division, MRC Environmental Epidemiology Unit, School
of Medicine, Southampton University Hospital, Tremona Road, Southampton, UK Elaine Dennison, Holly E Syddall, David I W Phillips & Cyrus Cooper Authors * Santiago Rodríguez View author
publications You can also search for this author inPubMed Google Scholar * Tom R Gaunt View author publications You can also search for this author inPubMed Google Scholar * Elaine Dennison
View author publications You can also search for this author inPubMed Google Scholar * Xiao-he Chen View author publications You can also search for this author inPubMed Google Scholar *
Holly E Syddall View author publications You can also search for this author inPubMed Google Scholar * David I W Phillips View author publications You can also search for this author
inPubMed Google Scholar * Cyrus Cooper View author publications You can also search for this author inPubMed Google Scholar * Ian N M Day View author publications You can also search for
this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Ian N M Day. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Rodríguez, S.,
Gaunt, T., Dennison, E. _et al._ Replication of _IGF2-INS-TH_*5 haplotype effect on obesity in older men and study of related phenotypes. _Eur J Hum Genet_ 14, 109–116 (2006).
https://doi.org/10.1038/sj.ejhg.5201505 Download citation * Received: 06 January 2005 * Revised: 29 July 2005 * Accepted: 16 September 2005 * Published: 26 October 2005 * Issue Date: January
2006 * DOI: https://doi.org/10.1038/sj.ejhg.5201505 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link
is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative KEYWORDS * obesity * _IGF2-INS-TH_ genomic region *
genetic association * replication * haplotype