Play all audios:
_Proc. Natl. Acad. Sci. USA_ 113, E4460–E4466 (2016) Credit: _PNAS_ In eukaryotes, the asymmetry of the plasma membrane is critical to its proper function, and this asymmetry is maintained
by phospholipid flippase enzymes that selectively transport phospholipids from the exofacial to the cytofacial leaflet of the membrane. In yeast, the P4-ATPase Dnf1 is a phosphatidylcholine
(PC) flippase and does not transport sphingomyelin (SM), even though the two have the same choline headgroup and highly similar fatty acyl chains. To understand how Dnf1 exerts such
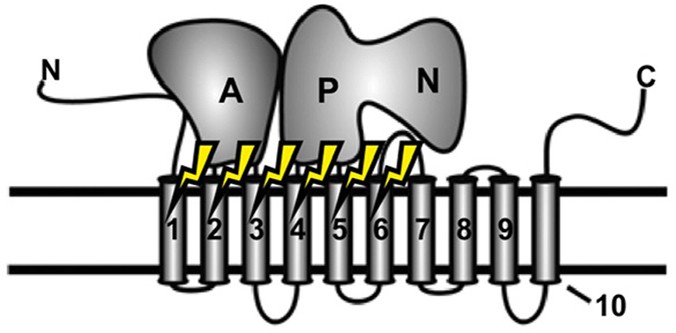
selectivity for its phospholipid substrates, Roland _et al_. used a directed evolution library of Dnf1 variants in a screen with fluorescently labeled PC and SM to identify flippase
sequences capable of selecting for SM instead of PC. A single amino acid change at the cytofacial side of the transmembrane domain enabled Dnf1 to select for the sphingosine backbone of SM
over the glycerol backbone of PC. Homology modeling and additional mutagenesis further elucidated how this position, along with others nearby in the exit-gate region of the protein,
discriminates between various phospholipid backbones as well as different headgroups. The insights into selectivity afforded by this study suggest how substrate phospholipids are coordinated
by the exit gate and should enable the design of new substrate-specific flippases for future studies of membrane asymmetry. Authors * Caitlin Deane View author publications You can also
search for this author inPubMed Google Scholar RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Deane, C. How to flip a flippase. _Nat Chem Biol_ 12, 763
(2016). https://doi.org/10.1038/nchembio.2199 Download citation * Published: 20 September 2016 * Issue Date: October 2016 * DOI: https://doi.org/10.1038/nchembio.2199 SHARE THIS ARTICLE
Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided
by the Springer Nature SharedIt content-sharing initiative